Corey Arnold
CytoFM: The first cytology foundation model
Apr 18, 2025Abstract:Cytology is essential for cancer diagnostics and screening due to its minimally invasive nature. However, the development of robust deep learning models for digital cytology is challenging due to the heterogeneity in staining and preparation methods of samples, differences across organs, and the limited availability of large, diverse, annotated datasets. Developing a task-specific model for every cytology application is impractical and non-cytology-specific foundation models struggle to generalize to tasks in this domain where the emphasis is on cell morphology. To address these challenges, we introduce CytoFM, the first cytology self-supervised foundation model. Using iBOT, a self-supervised Vision Transformer (ViT) training framework incorporating masked image modeling and self-distillation, we pretrain CytoFM on a diverse collection of cytology datasets to learn robust, transferable representations. We evaluate CytoFM on multiple downstream cytology tasks, including breast cancer classification and cell type identification, using an attention-based multiple instance learning framework. Our results demonstrate that CytoFM performs better on two out of three downstream tasks than existing foundation models pretrained on histopathology (UNI) or natural images (iBOT-Imagenet). Visualizations of learned representations demonstrate our model is able to attend to cytologically relevant features. Despite a small pre-training dataset, CytoFM's promising results highlight the ability of task-agnostic pre-training approaches to learn robust and generalizable features from cytology data.
Prototype-Guided Diffusion for Digital Pathology: Achieving Foundation Model Performance with Minimal Clinical Data
Apr 15, 2025



Abstract:Foundation models in digital pathology use massive datasets to learn useful compact feature representations of complex histology images. However, there is limited transparency into what drives the correlation between dataset size and performance, raising the question of whether simply adding more data to increase performance is always necessary. In this study, we propose a prototype-guided diffusion model to generate high-fidelity synthetic pathology data at scale, enabling large-scale self-supervised learning and reducing reliance on real patient samples while preserving downstream performance. Using guidance from histological prototypes during sampling, our approach ensures biologically and diagnostically meaningful variations in the generated data. We demonstrate that self-supervised features trained on our synthetic dataset achieve competitive performance despite using ~60x-760x less data than models trained on large real-world datasets. Notably, models trained using our synthetic data showed statistically comparable or better performance across multiple evaluation metrics and tasks, even when compared to models trained on orders of magnitude larger datasets. Our hybrid approach, combining synthetic and real data, further enhanced performance, achieving top results in several evaluations. These findings underscore the potential of generative AI to create compelling training data for digital pathology, significantly reducing the reliance on extensive clinical datasets and highlighting the efficiency of our approach.
Evaluation Of P300 Speller Performance Using Large Language Models Along With Cross-Subject Training
Oct 19, 2024



Abstract:Amyotrophic lateral sclerosis (ALS), a progressive neuromuscular degenerative disease, severely restricts patient communication capacity within a few years of onset, resulting in a significant deterioration of quality of life. The P300 speller brain computer interface (BCI) offers an alternative communication medium by leveraging a subject's EEG response to characters traditionally highlighted on a character grid on a graphical user interface (GUI). A recurring theme in P300-based research is enhancing performance to enable faster subject interaction. This study builds on that theme by addressing key limitations, particularly in the training of multi-subject classifiers, and by integrating advanced language models to optimize stimuli presentation and word prediction, thereby improving communication efficiency. Furthermore, various advanced large language models such as Generative Pre-Trained Transformer (GPT2), BERT, and BART, alongside Dijkstra's algorithm, are utilized to optimize stimuli and provide word completion choices based on the spelling history. In addition, a multi-layered smoothing approach is applied to allow for out-of-vocabulary (OOV) words. By conducting extensive simulations based on randomly sampled EEG data from subjects, we show substantial speed improvements in typing passages that include rare and out-of-vocabulary (OOV) words, with the extent of improvement varying depending on the language model utilized. The gains through such character-level interface optimizations are approximately 10%, and GPT2 for multi-word prediction provides gains of around 40%. In particular, some large language models achieve performance levels within 10% of the theoretical performance limits established in this study. In addition, both within and across subjects, training techniques are explored, and speed improvements are shown to hold in both cases.
High Performance P300 Spellers Using GPT2 Word Prediction With Cross-Subject Training
May 22, 2024Abstract:Amyotrophic lateral sclerosis (ALS) severely impairs patients' ability to communicate, often leading to a decline in their quality of life within a few years of diagnosis. The P300 speller brain-computer interface (BCI) offers an alternative communication method by interpreting a subject's EEG response to characters presented on a grid interface. This paper addresses the common speed limitations encountered in training efficient P300-based multi-subject classifiers by introducing innovative "across-subject" classifiers. We leverage a combination of the second-generation Generative Pre-Trained Transformer (GPT2) and Dijkstra's algorithm to optimize stimuli and suggest word completion choices based on typing history. Additionally, we employ a multi-layered smoothing technique to accommodate out-of-vocabulary (OOV) words. Through extensive simulations involving random sampling of EEG data from subjects, we demonstrate significant speed enhancements in typing passages containing rare and OOV words. These optimizations result in approximately 10% improvement in character-level typing speed and up to 40% improvement in multi-word prediction. We demonstrate that augmenting standard row/column highlighting techniques with layered word prediction yields close-to-optimal performance. Furthermore, we explore both "within-subject" and "across-subject" training techniques, showing that speed improvements are consistent across both approaches.
Multi-site, Multi-domain Airway Tree Modeling : A Public Benchmark for Pulmonary Airway Segmentation
Mar 10, 2023



Abstract:Open international challenges are becoming the de facto standard for assessing computer vision and image analysis algorithms. In recent years, new methods have extended the reach of pulmonary airway segmentation that is closer to the limit of image resolution. Since EXACT'09 pulmonary airway segmentation, limited effort has been directed to quantitative comparison of newly emerged algorithms driven by the maturity of deep learning based approaches and clinical drive for resolving finer details of distal airways for early intervention of pulmonary diseases. Thus far, public annotated datasets are extremely limited, hindering the development of data-driven methods and detailed performance evaluation of new algorithms. To provide a benchmark for the medical imaging community, we organized the Multi-site, Multi-domain Airway Tree Modeling (ATM'22), which was held as an official challenge event during the MICCAI 2022 conference. ATM'22 provides large-scale CT scans with detailed pulmonary airway annotation, including 500 CT scans (300 for training, 50 for validation, and 150 for testing). The dataset was collected from different sites and it further included a portion of noisy COVID-19 CTs with ground-glass opacity and consolidation. Twenty-three teams participated in the entire phase of the challenge and the algorithms for the top ten teams are reviewed in this paper. Quantitative and qualitative results revealed that deep learning models embedded with the topological continuity enhancement achieved superior performance in general. ATM'22 challenge holds as an open-call design, the training data and the gold standard evaluation are available upon successful registration via its homepage.
Semi-supervised learning based on generative adversarial network: a comparison between good GAN and bad GAN approach
May 17, 2019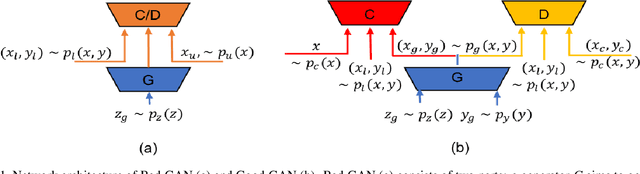
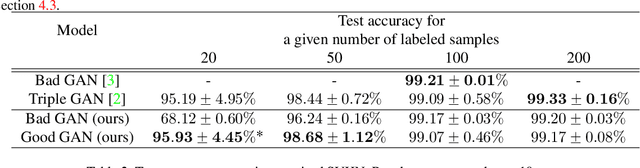
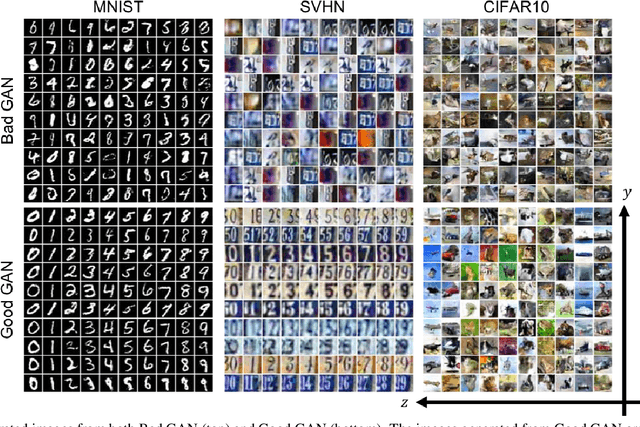

Abstract:Recently, semi-supervised learning methods based on generative adversarial networks (GANs) have received much attention. Among them, two distinct approaches have achieved competitive results on a variety of benchmark datasets. Bad GAN learns a classifier with unrealistic samples distributed on the complement of the support of the input data. Conversely, Triple GAN consists of a three-player game that tries to leverage good generated samples to boost classification results. In this paper, we perform a comprehensive comparison of these two approaches on different benchmark datasets. We demonstrate their different properties on image generation, and sensitivity to the amount of labeled data provided. By comprehensively comparing these two methods, we hope to shed light on the future of GAN-based semi-supervised learning.
Semi-supervised Rare Disease Detection Using Generative Adversarial Network
Dec 03, 2018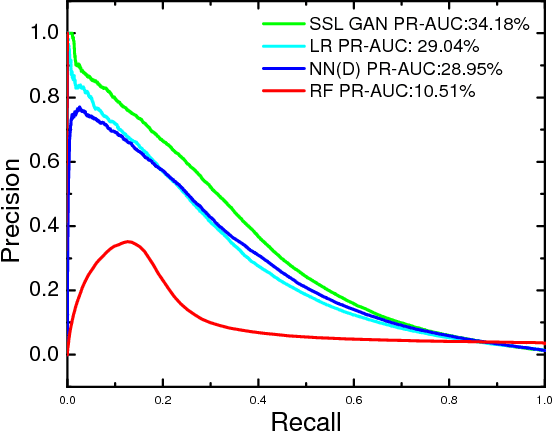
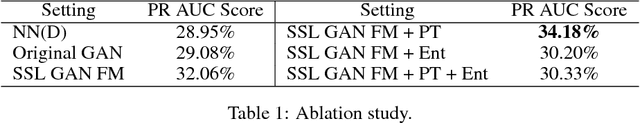
Abstract:Rare diseases affect a relatively small number of people, which limits investment in research for treatments and cures. Developing an efficient method for rare disease detection is a crucial first step towards subsequent clinical research. In this paper, we present a semi-supervised learning framework for rare disease detection using generative adversarial networks. Our method takes advantage of the large amount of unlabeled data for disease detection and achieves the best results in terms of precision-recall score compared to baseline techniques.
Source-LDA: Enhancing probabilistic topic models using prior knowledge sources
May 17, 2017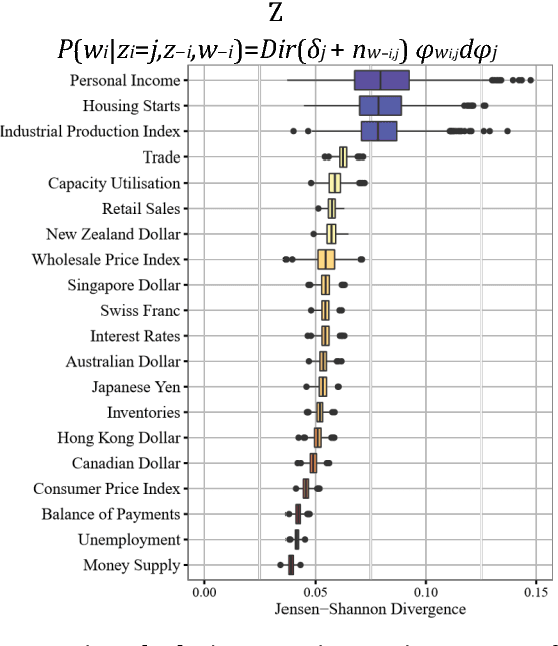
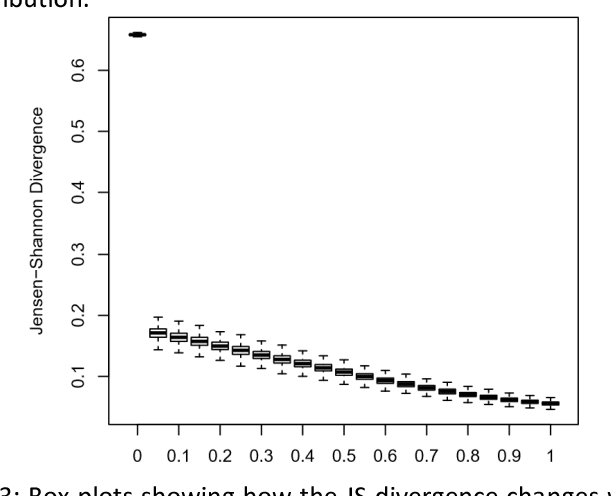
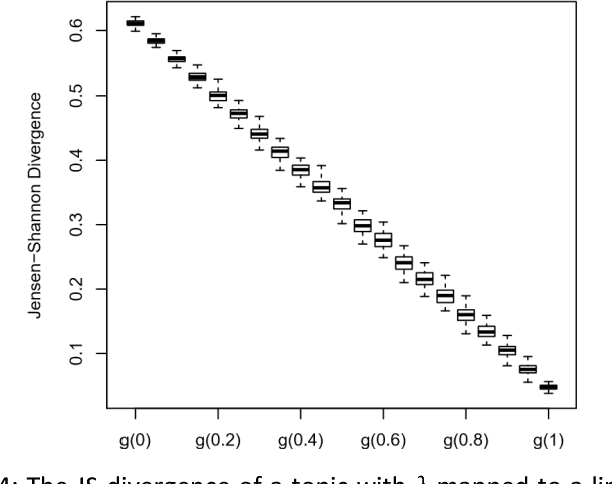
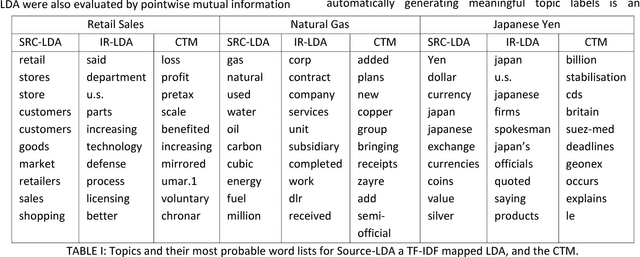
Abstract:A popular approach to topic modeling involves extracting co-occurring n-grams of a corpus into semantic themes. The set of n-grams in a theme represents an underlying topic, but most topic modeling approaches are not able to label these sets of words with a single n-gram. Such labels are useful for topic identification in summarization systems. This paper introduces a novel approach to labeling a group of n-grams comprising an individual topic. The approach taken is to complement the existing topic distributions over words with a known distribution based on a predefined set of topics. This is done by integrating existing labeled knowledge sources representing known potential topics into the probabilistic topic model. These knowledge sources are translated into a distribution and used to set the hyperparameters of the Dirichlet generated distribution over words. In the inference these modified distributions guide the convergence of the latent topics to conform with the complementary distributions. This approach ensures that the topic inference process is consistent with existing knowledge. The label assignment from the complementary knowledge sources are then transferred to the latent topics of the corpus. The results show both accurate label assignment to topics as well as improved topic generation than those obtained using various labeling approaches based off Latent Dirichlet allocation (LDA).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge