Cheng Jiang
Tencent AI Lab, Shenzhen, China
A self-supervised framework for learning whole slide representations
Feb 09, 2024



Abstract:Whole slide imaging is fundamental to biomedical microscopy and computational pathology. However, whole slide images (WSIs) present a complex computer vision challenge due to their gigapixel size, diverse histopathologic features, spatial heterogeneity, and limited/absent data annotations. These challenges highlight that supervised training alone can result in suboptimal whole slide representations. Self-supervised representation learning can achieve high-quality WSI visual feature learning for downstream diagnostic tasks, such as cancer diagnosis or molecular genetic prediction. Here, we present a general self-supervised whole slide learning (S3L) framework for gigapixel-scale self-supervision of WSIs. S3L combines data transformation strategies from transformer-based vision and language modeling into a single unified framework to generate paired views for self-supervision. S3L leverages the inherent regional heterogeneity, histologic feature variability, and information redundancy within WSIs to learn high-quality whole-slide representations. We benchmark S3L visual representations on two diagnostic tasks for two biomedical microscopy modalities. S3L significantly outperforms WSI baselines for cancer diagnosis and genetic mutation prediction. Additionally, S3L achieves good performance using both in-domain and out-of-distribution patch encoders, demonstrating good flexibility and generalizability.
Development and validation of an artificial intelligence model to accurately predict spinopelvic parameters
Feb 09, 2024Abstract:Objective. Achieving appropriate spinopelvic alignment has been shown to be associated with improved clinical symptoms. However, measurement of spinopelvic radiographic parameters is time-intensive and interobserver reliability is a concern. Automated measurement tools have the promise of rapid and consistent measurements, but existing tools are still limited by some degree of manual user-entry requirements. This study presents a novel artificial intelligence (AI) tool called SpinePose that automatically predicts spinopelvic parameters with high accuracy without the need for manual entry. Methods. SpinePose was trained and validated on 761 sagittal whole-spine X-rays to predict sagittal vertical axis (SVA), pelvic tilt (PT), pelvic incidence (PI), sacral slope (SS), lumbar lordosis (LL), T1-pelvic angle (T1PA), and L1-pelvic angle (L1PA). A separate test set of 40 X-rays was labeled by 4 reviewers, including fellowship-trained spine surgeons and a fellowship-trained radiologist with neuroradiology subspecialty certification. Median errors relative to the most senior reviewer were calculated to determine model accuracy on test images. Intraclass correlation coefficients (ICC) were used to assess inter-rater reliability. Results. SpinePose exhibited the following median (interquartile range) parameter errors: SVA: 2.2(2.3)mm, p=0.93; PT: 1.3(1.2){\deg}, p=0.48; SS: 1.7(2.2){\deg}, p=0.64; PI: 2.2(2.1){\deg}, p=0.24; LL: 2.6(4.0){\deg}, p=0.89; T1PA: 1.1(0.9){\deg}, p=0.42; and L1PA: 1.4(1.6){\deg}, p=0.49. Model predictions also exhibited excellent reliability at all parameters (ICC: 0.91-1.0). Conclusions. SpinePose accurately predicted spinopelvic parameters with excellent reliability comparable to fellowship-trained spine surgeons and neuroradiologists. Utilization of predictive AI tools in spinal imaging can substantially aid in patient selection and surgical planning.
Artificial-intelligence-based molecular classification of diffuse gliomas using rapid, label-free optical imaging
Mar 23, 2023Abstract:Molecular classification has transformed the management of brain tumors by enabling more accurate prognostication and personalized treatment. However, timely molecular diagnostic testing for patients with brain tumors is limited, complicating surgical and adjuvant treatment and obstructing clinical trial enrollment. In this study, we developed DeepGlioma, a rapid ($< 90$ seconds), artificial-intelligence-based diagnostic screening system to streamline the molecular diagnosis of diffuse gliomas. DeepGlioma is trained using a multimodal dataset that includes stimulated Raman histology (SRH); a rapid, label-free, non-consumptive, optical imaging method; and large-scale, public genomic data. In a prospective, multicenter, international testing cohort of patients with diffuse glioma ($n=153$) who underwent real-time SRH imaging, we demonstrate that DeepGlioma can predict the molecular alterations used by the World Health Organization to define the adult-type diffuse glioma taxonomy (IDH mutation, 1p19q co-deletion and ATRX mutation), achieving a mean molecular classification accuracy of $93.3\pm 1.6\%$. Our results represent how artificial intelligence and optical histology can be used to provide a rapid and scalable adjunct to wet lab methods for the molecular screening of patients with diffuse glioma.
Hierarchical discriminative learning improves visual representations of biomedical microscopy
Mar 02, 2023



Abstract:Learning high-quality, self-supervised, visual representations is essential to advance the role of computer vision in biomedical microscopy and clinical medicine. Previous work has focused on self-supervised representation learning (SSL) methods developed for instance discrimination and applied them directly to image patches, or fields-of-view, sampled from gigapixel whole-slide images (WSIs) used for cancer diagnosis. However, this strategy is limited because it (1) assumes patches from the same patient are independent, (2) neglects the patient-slide-patch hierarchy of clinical biomedical microscopy, and (3) requires strong data augmentations that can degrade downstream performance. Importantly, sampled patches from WSIs of a patient's tumor are a diverse set of image examples that capture the same underlying cancer diagnosis. This motivated HiDisc, a data-driven method that leverages the inherent patient-slide-patch hierarchy of clinical biomedical microscopy to define a hierarchical discriminative learning task that implicitly learns features of the underlying diagnosis. HiDisc uses a self-supervised contrastive learning framework in which positive patch pairs are defined based on a common ancestry in the data hierarchy, and a unified patch, slide, and patient discriminative learning objective is used for visual SSL. We benchmark HiDisc visual representations on two vision tasks using two biomedical microscopy datasets, and demonstrate that (1) HiDisc pretraining outperforms current state-of-the-art self-supervised pretraining methods for cancer diagnosis and genetic mutation prediction, and (2) HiDisc learns high-quality visual representations using natural patch diversity without strong data augmentations.
MixFormer: End-to-End Tracking with Iterative Mixed Attention
Feb 09, 2023



Abstract:Visual object tracking often employs a multi-stage pipeline of feature extraction, target information integration, and bounding box estimation. To simplify this pipeline and unify the process of feature extraction and target information integration, in this paper, we present a compact tracking framework, termed as MixFormer, built upon transformers. Our core design is to utilize the flexibility of attention operations, and propose a Mixed Attention Module (MAM) for simultaneous feature extraction and target information integration. This synchronous modeling scheme allows to extract target-specific discriminative features and perform extensive communication between target and search area. Based on MAM, we build our MixFormer trackers simply by stacking multiple MAMs and placing a localization head on top. Specifically, we instantiate two types of MixFormer trackers, a hierarchical tracker MixCvT, and a non-hierarchical tracker MixViT. For these two trackers, we investigate a series of pre-training methods and uncover the different behaviors between supervised pre-training and self-supervised pre-training in our MixFormer trackers. We also extend the masked pre-training to our MixFormer trackers and design the competitive TrackMAE pre-training technique. Finally, to handle multiple target templates during online tracking, we devise an asymmetric attention scheme in MAM to reduce computational cost, and propose an effective score prediction module to select high-quality templates. Our MixFormer trackers set a new state-of-the-art performance on seven tracking benchmarks, including LaSOT, TrackingNet, VOT2020, GOT-10k, OTB100 and UAV123. In particular, our MixViT-L achieves AUC score of 73.3% on LaSOT, 86.1% on TrackingNet, EAO of 0.584 on VOT2020, and AO of 75.7% on GOT-10k. Code and trained models are publicly available at https://github.com/MCG-NJU/MixFormer.
OpenSRH: optimizing brain tumor surgery using intraoperative stimulated Raman histology
Jun 16, 2022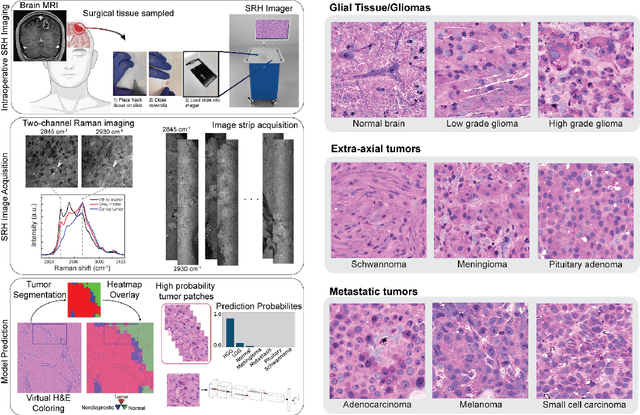
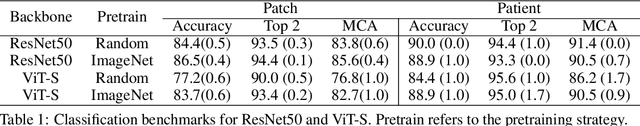
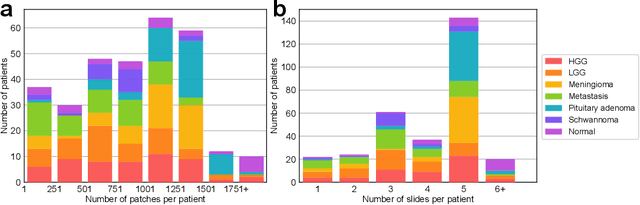
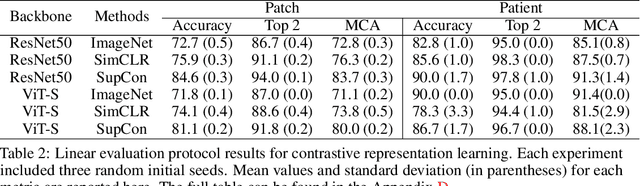
Abstract:Accurate intraoperative diagnosis is essential for providing safe and effective care during brain tumor surgery. Our standard-of-care diagnostic methods are time, resource, and labor intensive, which restricts access to optimal surgical treatments. To address these limitations, we propose an alternative workflow that combines stimulated Raman histology (SRH), a rapid optical imaging method, with deep learning-based automated interpretation of SRH images for intraoperative brain tumor diagnosis and real-time surgical decision support. Here, we present OpenSRH, the first public dataset of clinical SRH images from 300+ brain tumors patients and 1300+ unique whole slide optical images. OpenSRH contains data from the most common brain tumors diagnoses, full pathologic annotations, whole slide tumor segmentations, raw and processed optical imaging data for end-to-end model development and validation. We provide a framework for patch-based whole slide SRH classification and inference using weak (i.e. patient-level) diagnostic labels. Finally, we benchmark two computer vision tasks: multiclass histologic brain tumor classification and patch-based contrastive representation learning. We hope OpenSRH will facilitate the clinical translation of rapid optical imaging and real-time ML-based surgical decision support in order to improve the access, safety, and efficacy of cancer surgery in the era of precision medicine. Dataset access, code, and benchmarks are available at opensrh.mlins.org.
3D Shuffle-Mixer: An Efficient Context-Aware Vision Learner of Transformer-MLP Paradigm for Dense Prediction in Medical Volume
Apr 14, 2022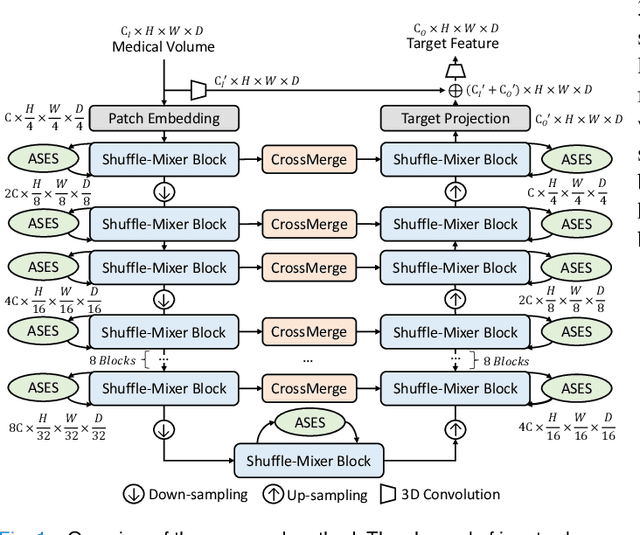
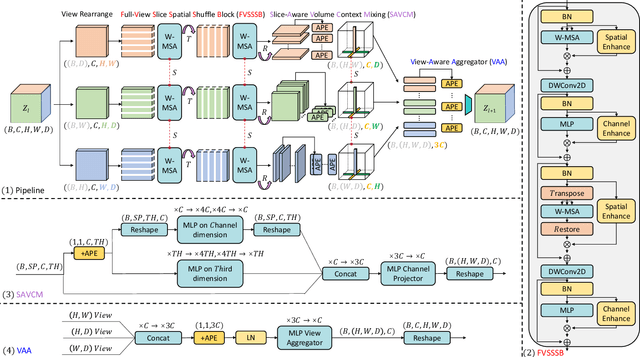
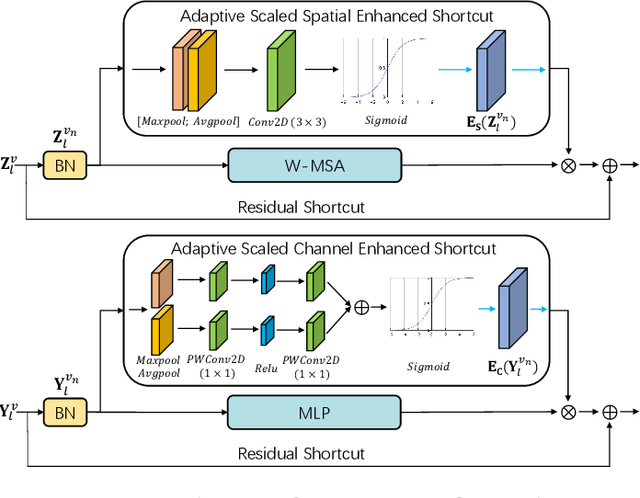
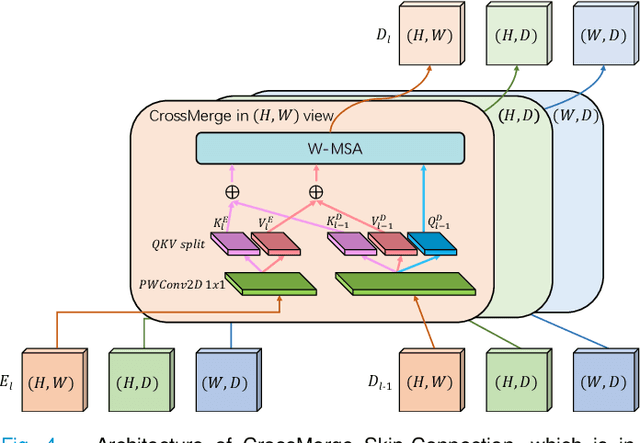
Abstract:Dense prediction in medical volume provides enriched guidance for clinical analysis. CNN backbones have met bottleneck due to lack of long-range dependencies and global context modeling power. Recent works proposed to combine vision transformer with CNN, due to its strong global capture ability and learning capability. However, most works are limited to simply applying pure transformer with several fatal flaws (i.e., lack of inductive bias, heavy computation and little consideration for 3D data). Therefore, designing an elegant and efficient vision transformer learner for dense prediction in medical volume is promising and challenging. In this paper, we propose a novel 3D Shuffle-Mixer network of a new Local Vision Transformer-MLP paradigm for medical dense prediction. In our network, a local vision transformer block is utilized to shuffle and learn spatial context from full-view slices of rearranged volume, a residual axial-MLP is designed to mix and capture remaining volume context in a slice-aware manner, and a MLP view aggregator is employed to project the learned full-view rich context to the volume feature in a view-aware manner. Moreover, an Adaptive Scaled Enhanced Shortcut is proposed for local vision transformer to enhance feature along spatial and channel dimensions adaptively, and a CrossMerge is proposed to skip-connects the multi-scale feature appropriately in the pyramid architecture. Extensive experiments demonstrate the proposed model outperforms other state-of-the-art medical dense prediction methods.
Fusion of medical imaging and electronic health records with attention and multi-head machanisms
Dec 22, 2021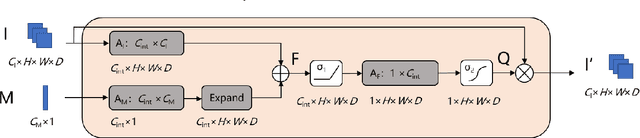
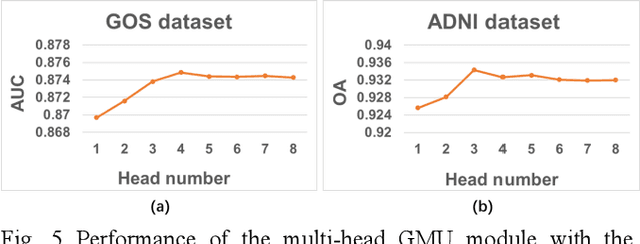
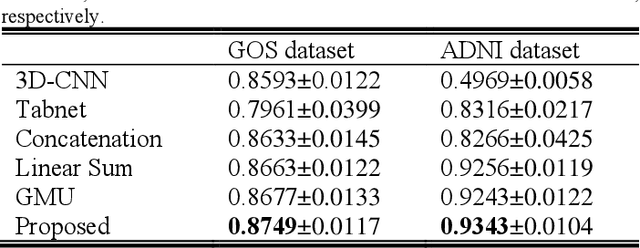

Abstract:Doctors often make diagonostic decisions based on patient's image scans, such as magnetic resonance imaging (MRI), and patient's electronic health records (EHR) such as age, gender, blood pressure and so on. Despite a lot of automatic methods have been proposed for either image or text analysis in computer vision or natural language research areas, much fewer studies have been developed for the fusion of medical image and EHR data for medical problems. Among existing early or intermediate fusion methods, concatenation of features from both modalities is still a mainstream. For a better exploiting of image and EHR data, we propose a multi-modal attention module which use EHR data to help the selection of important regions during image feature extraction process conducted by traditional CNN. Moreover, we propose to incorporate multi-head machnism to gated multimodal unit (GMU) to make it able to parallelly fuse image and EHR features in different subspaces. With the help of the two modules, existing CNN architecture can be enhanced using both modalities. Experiments on predicting Glasgow outcome scale (GOS) of intracerebral hemorrhage patients and classifying Alzheimer's Disease showed the proposed method can automatically focus on task-related areas and achieve better results by making better use of image and EHR features.
Two-stream Convolutional Networks for Multi-frame Face Anti-spoofing
Aug 09, 2021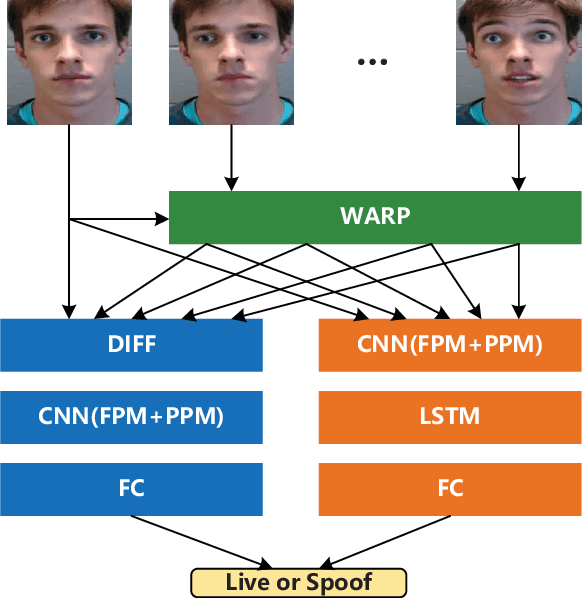
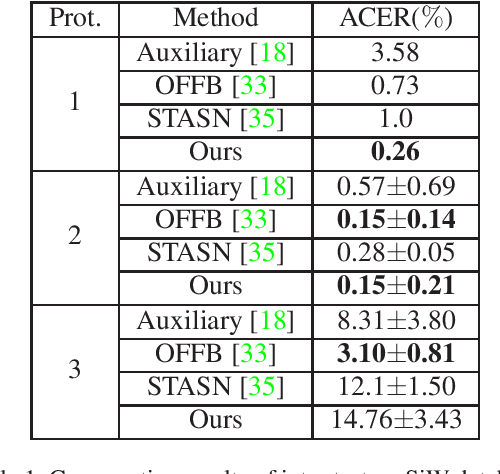
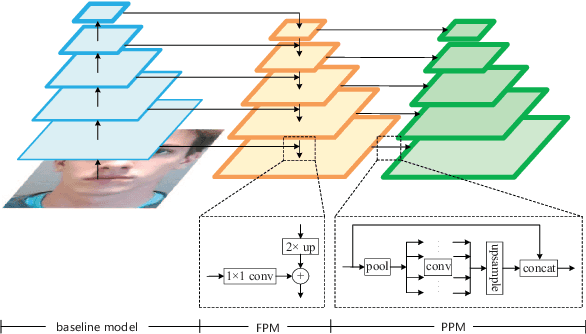
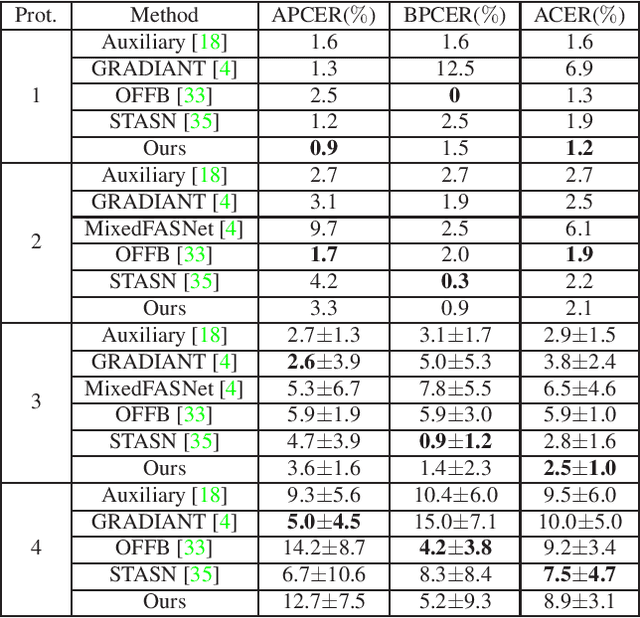
Abstract:Face anti-spoofing is an important task to protect the security of face recognition. Most of previous work either struggle to capture discriminative and generalizable feature or rely on auxiliary information which is unavailable for most of industrial product. Inspired by the video classification work, we propose an efficient two-stream model to capture the key differences between live and spoof faces, which takes multi-frames and RGB difference as input respectively. Feature pyramid modules with two opposite fusion directions and pyramid pooling modules are applied to enhance feature representation. We evaluate the proposed method on the datasets of Siw, Oulu-NPU, CASIA-MFSD and Replay-Attack. The results show that our model achieves the state-of-the-art results on most of datasets' protocol with much less parameter size.
Contrastive Representation Learning for Rapid Intraoperative Diagnosis of Skull Base Tumors Imaged Using Stimulated Raman Histology
Aug 08, 2021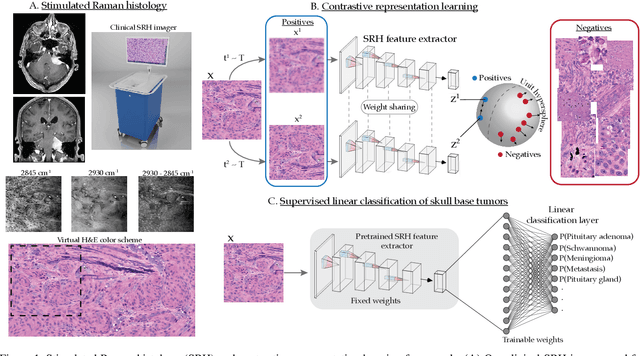
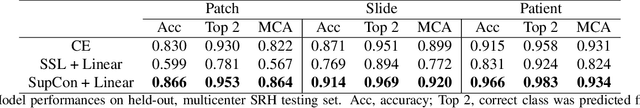
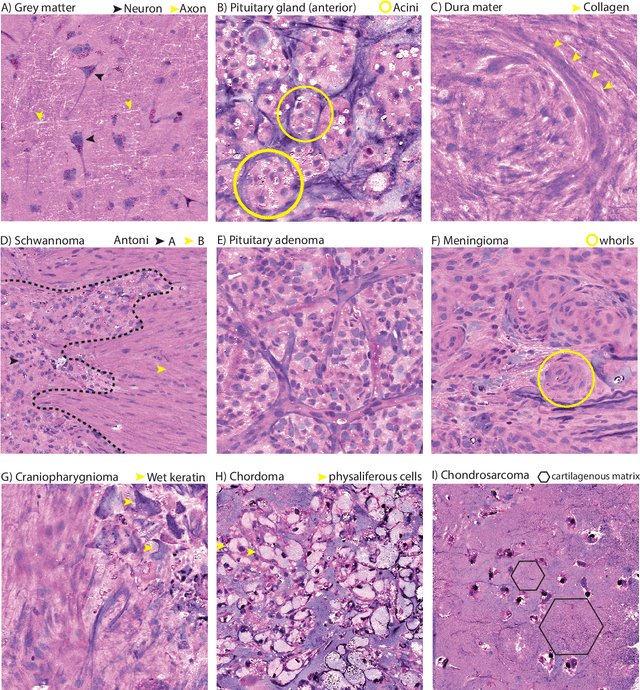
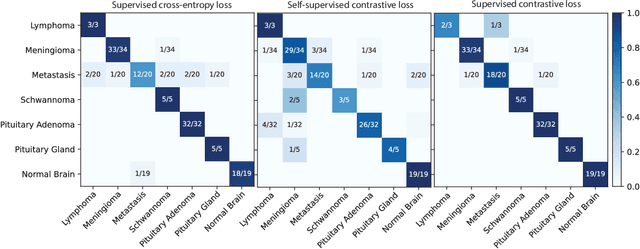
Abstract:Background: Accurate diagnosis of skull base tumors is essential for providing personalized surgical treatment strategies. Intraoperative diagnosis can be challenging due to tumor diversity and lack of intraoperative pathology resources. Objective: To develop an independent and parallel intraoperative pathology workflow that can provide rapid and accurate skull base tumor diagnoses using label-free optical imaging and artificial intelligence (AI). Method: We used a fiber laser-based, label-free, non-consumptive, high-resolution microscopy method ($<$ 60 sec per 1 $\times$ 1 mm$^\text{2}$), called stimulated Raman histology (SRH), to image a consecutive, multicenter cohort of skull base tumor patients. SRH images were then used to train a convolutional neural network (CNN) model using three representation learning strategies: cross-entropy, self-supervised contrastive learning, and supervised contrastive learning. Our trained CNN models were tested on a held-out, multicenter SRH dataset. Results: SRH was able to image the diagnostic features of both benign and malignant skull base tumors. Of the three representation learning strategies, supervised contrastive learning most effectively learned the distinctive and diagnostic SRH image features for each of the skull base tumor types. In our multicenter testing set, cross-entropy achieved an overall diagnostic accuracy of 91.5%, self-supervised contrastive learning 83.9%, and supervised contrastive learning 96.6%. Our trained model was able to identify tumor-normal margins and detect regions of microscopic tumor infiltration in whole-slide SRH images. Conclusion: SRH with AI models trained using contrastive representation learning can provide rapid and accurate intraoperative diagnosis of skull base tumors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge