Petru Manescu
MORPHFED: Federated Learning for Cross-institutional Blood Morphology Analysis
Jan 07, 2026Abstract:Automated blood morphology analysis can support hematological diagnostics in low- and middle-income countries (LMICs) but remains sensitive to dataset shifts from staining variability, imaging differences, and rare morphologies. Building centralized datasets to capture this diversity is often infeasible due to privacy regulations and data-sharing restrictions. We introduce a federated learning framework for white blood cell morphology analysis that enables collaborative training across institutions without exchanging training data. Using blood films from multiple clinical sites, our federated models learn robust, domain-invariant representations while preserving complete data privacy. Evaluations across convolutional and transformer-based architectures show that federated training achieves strong cross-site performance and improved generalization to unseen institutions compared to centralized training. These findings highlight federated learning as a practical and privacy-preserving approach for developing equitable, scalable, and generalizable medical imaging AI in resource-limited healthcare environments.
HemBLIP: A Vision-Language Model for Interpretable Leukemia Cell Morphology Analysis
Jan 07, 2026Abstract:Microscopic evaluation of white blood cell morphology is central to leukemia diagnosis, yet current deep learning models often act as black boxes, limiting clinical trust and adoption. We introduce HemBLIP, a vision language model designed to generate interpretable, morphology aware descriptions of peripheral blood cells. Using a newly constructed dataset of 14k healthy and leukemic cells paired with expert-derived attribute captions, we adapt a general-purpose VLM via both full fine-tuning and LoRA based parameter efficient training, and benchmark against the biomedical foundation model MedGEMMA. HemBLIP achieves higher caption quality and morphological accuracy, while LoRA adaptation provides further gains with significantly reduced computational cost. These results highlight the promise of vision language models for transparent and scalable hematological diagnostics.
LesionTABE: Equitable AI for Skin Lesion Detection
Jan 06, 2026Abstract:Bias remains a major barrier to the clinical adoption of AI in dermatology, as diagnostic models underperform on darker skin tones. We present LesionTABE, a fairness-centric framework that couples adversarial debiasing with dermatology-specific foundation model embeddings. Evaluated across multiple datasets covering both malignant and inflammatory conditions, LesionTABE achieves over a 25\% improvement in fairness metrics compared to a ResNet-152 baseline, outperforming existing debiasing methods while simultaneously enhancing overall diagnostic accuracy. These results highlight the potential of foundation model debiasing as a step towards equitable clinical AI adoption.
Cognitive Maps in Language Models: A Mechanistic Analysis of Spatial Planning
Nov 17, 2025



Abstract:How do large language models solve spatial navigation tasks? We investigate this by training GPT-2 models on three spatial learning paradigms in grid environments: passive exploration (Foraging Model- predicting steps in random walks), goal-directed planning (generating optimal shortest paths) on structured Hamiltonian paths (SP-Hamiltonian), and a hybrid model fine-tuned with exploratory data (SP-Random Walk). Using behavioural, representational and mechanistic analyses, we uncover two fundamentally different learned algorithms. The Foraging model develops a robust, map-like representation of space, akin to a 'cognitive map'. Causal interventions reveal that it learns to consolidate spatial information into a self-sufficient coordinate system, evidenced by a sharp phase transition where its reliance on historical direction tokens vanishes by the middle layers of the network. The model also adopts an adaptive, hierarchical reasoning system, switching between a low-level heuristic for short contexts and map-based inference for longer ones. In contrast, the goal-directed models learn a path-dependent algorithm, remaining reliant on explicit directional inputs throughout all layers. The hybrid model, despite demonstrating improved generalisation over its parent, retains the same path-dependent strategy. These findings suggest that the nature of spatial intelligence in transformers may lie on a spectrum, ranging from generalisable world models shaped by exploratory data to heuristics optimised for goal-directed tasks. We provide a mechanistic account of this generalisation-optimisation trade-off and highlight how the choice of training regime influences the strategies that emerge.
Benchmarking Retinal Blood Vessel Segmentation Models for Cross-Dataset and Cross-Disease Generalization
Jun 21, 2024



Abstract:Retinal blood vessel segmentation can extract clinically relevant information from fundus images. As manual tracing is cumbersome, algorithms based on Convolution Neural Networks have been developed. Such studies have used small publicly available datasets for training and measuring performance, running the risk of overfitting. Here, we provide a rigorous benchmark for various architectural and training choices commonly used in the literature on the largest dataset published to date. We train and evaluate five published models on the publicly available FIVES fundus image dataset, which exceeds previous ones in size and quality and which contains also images from common ophthalmological conditions (diabetic retinopathy, age-related macular degeneration, glaucoma). We compare the performance of different model architectures across different loss functions, levels of image qualitiy and ophthalmological conditions and assess their ability to perform well in the face of disease-induced domain shifts. Given sufficient training data, basic architectures such as U-Net perform just as well as more advanced ones, and transfer across disease-induced domain shifts typically works well for most architectures. However, we find that image quality is a key factor determining segmentation outcomes. When optimizing for segmentation performance, investing into a well curated dataset to train a standard architecture yields better results than tuning a sophisticated architecture on a smaller dataset or one with lower image quality. We distilled the utility of architectural advances in terms of their clinical relevance therefore providing practical guidance for model choices depending on the circumstances of the clinical setting
Deep learning-based detection of morphological features associated with hypoxia in H&E breast cancer whole slide images
Nov 21, 2023


Abstract:Hypoxia occurs when tumour cells outgrow their blood supply, leading to regions of low oxygen levels within the tumour. Calculating hypoxia levels can be an important step in understanding the biology of tumours, their clinical progression and response to treatment. This study demonstrates a novel application of deep learning to evaluate hypoxia in the context of breast cancer histomorphology. More precisely, we show that Weakly Supervised Deep Learning (WSDL) models can accurately detect hypoxia associated features in routine Hematoxylin and Eosin (H&E) whole slide images (WSI). We trained and evaluated a deep Multiple Instance Learning model on tiles from WSI H&E tissue from breast cancer primary sites (n=240) obtaining on average an AUC of 0.87 on a left-out test set. We also showed significant differences between features of hypoxic and normoxic tissue regions as distinguished by the WSDL models. Such DL hypoxia H&E WSI detection models could potentially be extended to other tumour types and easily integrated into the pathology workflow without requiring additional costly assays.
Automated Detection of Acute Promyelocytic Leukemia in Blood Films and Bone Marrow Aspirates with Annotation-free Deep Learning
Mar 20, 2022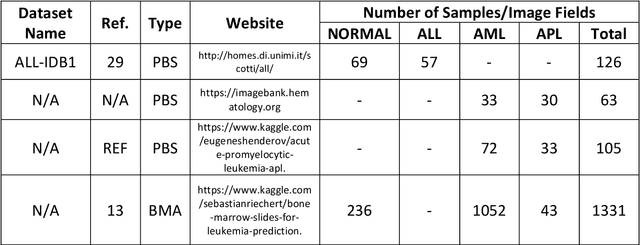
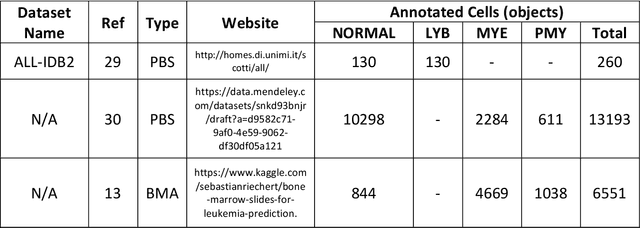
Abstract:While optical microscopy inspection of blood films and bone marrow aspirates by a hematologist is a crucial step in establishing diagnosis of acute leukemia, especially in low-resource settings where other diagnostic modalities might not be available, the task remains time-consuming and prone to human inconsistencies. This has an impact especially in cases of Acute Promyelocytic Leukemia (APL) that require urgent treatment. Integration of automated computational hematopathology into clinical workflows can improve the throughput of these services and reduce cognitive human error. However, a major bottleneck in deploying such systems is a lack of sufficient cell morphological object-labels annotations to train deep learning models. We overcome this by leveraging patient diagnostic labels to train weakly-supervised models that detect different types of acute leukemia. We introduce a deep learning approach, Multiple Instance Learning for Leukocyte Identification (MILLIE), able to perform automated reliable analysis of blood films with minimal supervision. Without being trained to classify individual cells, MILLIE differentiates between acute lymphoblastic and myeloblastic leukemia in blood films. More importantly, MILLIE detects APL in blood films (AUC 0.94+/-0.04) and in bone marrow aspirates (AUC 0.99+/-0.01). MILLIE is a viable solution to augment the throughput of clinical pathways that require assessment of blood film microscopy.
Data-Driven Malaria Prevalence Prediction in Large Densely-Populated Urban Holoendemic sub-Saharan West Africa: Harnessing Machine Learning Approaches and 22-years of Prospectively Collected Data
Jun 18, 2019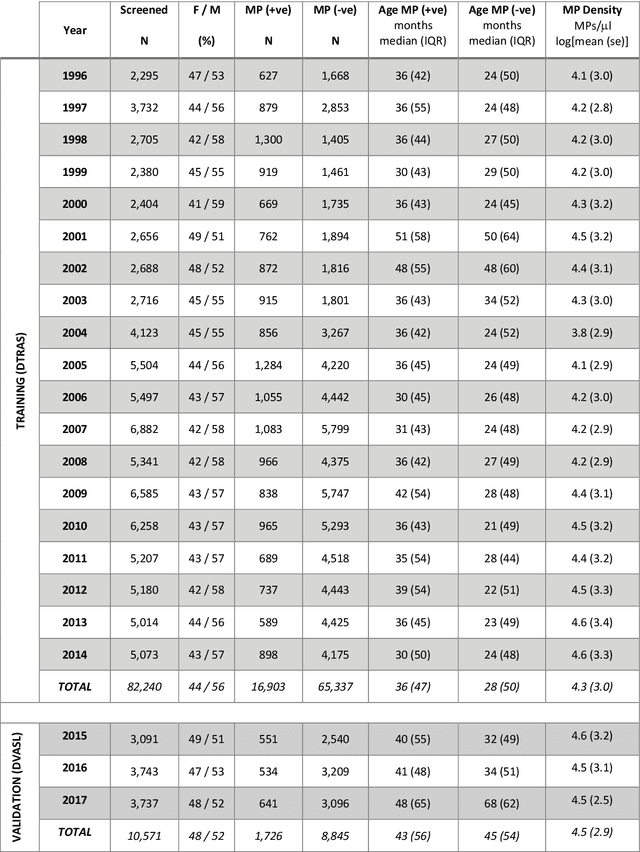
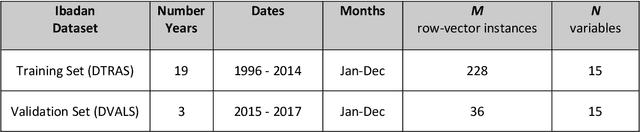
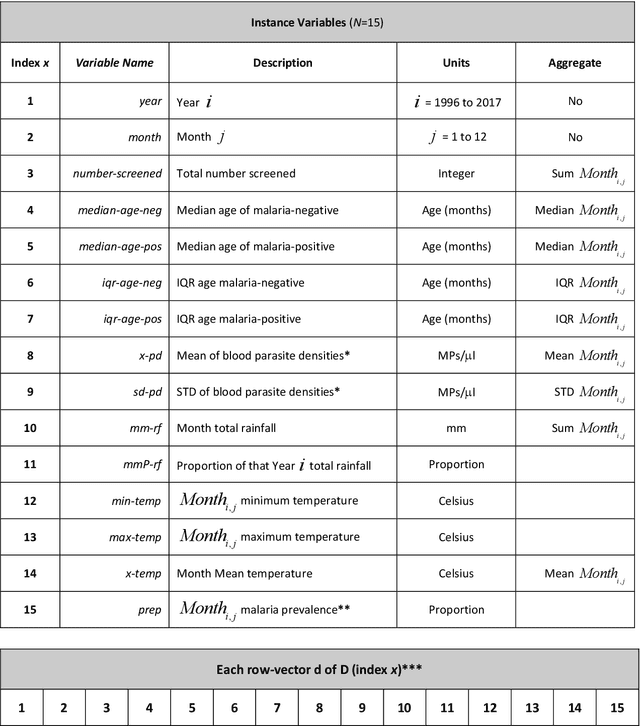
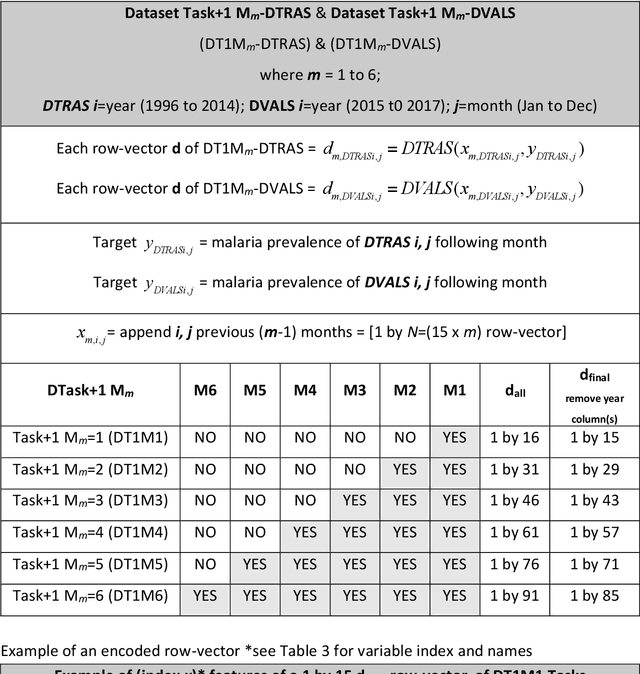
Abstract:Plasmodium falciparum malaria still poses one of the greatest threats to human life with over 200 million cases globally leading to half-million deaths annually. Of these, 90% of cases and of the mortality occurs in sub-Saharan Africa, mostly among children. Although malaria prediction systems are central to the 2016-2030 malaria Global Technical Strategy, currently these are inadequate at capturing and estimating the burden of disease in highly endemic countries. We developed and validated a computational system that exploits the predictive power of current Machine Learning approaches on 22-years of prospective data from the high-transmission holoendemic malaria urban-densely-populated sub-Saharan West-Africa metropolis of Ibadan. Our dataset of >9x104 screened study participants attending our clinical and community services from 1996 to 2017 contains monthly prevalence, temporal, environmental and host features. Our Locality-specific Elastic-Net based Malaria Prediction System (LEMPS) achieves good generalization performance, both in magnitude and direction of the prediction, when tasked to predict monthly prevalence on previously unseen validation data (MAE<=6x10-2, MSE<=7x10-3) within a range of (+0.1 to -0.05) error-tolerance which is relevant and usable for aiding decision-support in a holoendemic setting. LEMPS is well-suited for malaria prediction, where there are multiple features which are correlated with one another, and trading-off between regularization-strength L1-norm and L2-norm allows the system to retain stability. Data-driven systems are critical for regionally-adaptable surveillance, management of control strategies and resource allocation across stretched healthcare systems.
Deep Learning Enhanced Extended Depth-of-Field for Thick Blood-Film Malaria High-Throughput Microscopy
Jun 18, 2019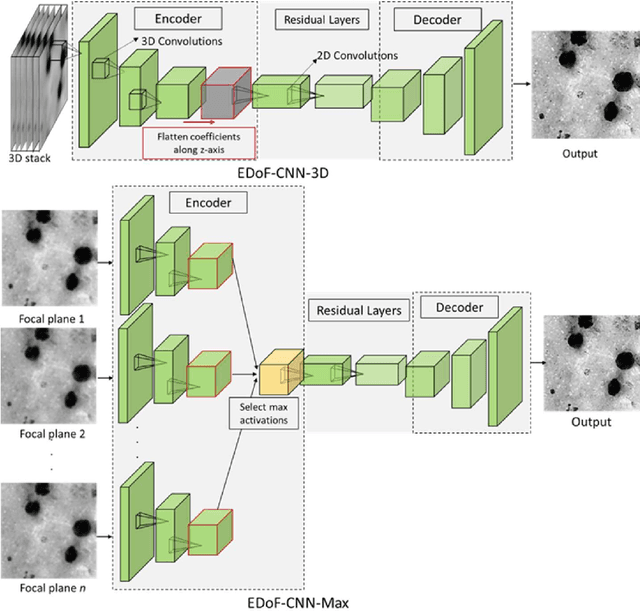
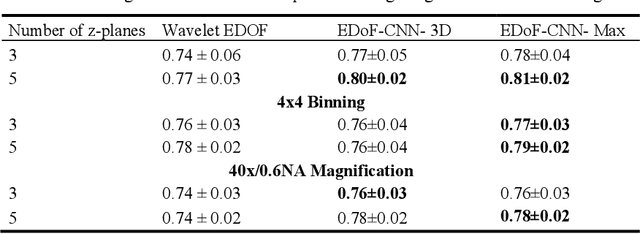
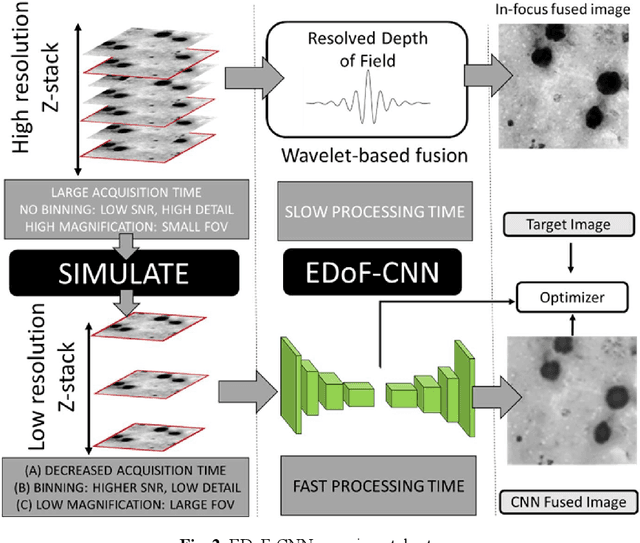
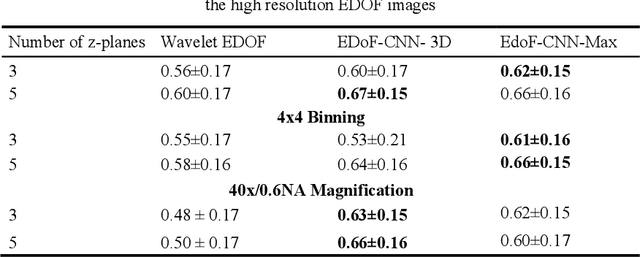
Abstract:Fast accurate diagnosis of malaria is still a global health challenge for which automated digital-pathology approaches could provide scalable solutions amenable to be deployed in low-to-middle income countries. Here we address the problem of Extended Depth-of-Field (EDoF) in thick blood film microscopy for rapid automated malaria diagnosis. High magnification oil-objectives (100x) with large numerical aperture are usually preferred to resolve the fine structural details that help separate true parasites from distractors. However, such objectives have a very limited depth-of-field requiring the acquisition of a series of images at different focal planes per field of view (FOV). Current EDoF techniques based on multi-scale decompositions are time consuming and therefore not suited for high-throughput analysis of specimens. To overcome this challenge, we developed a new deep learning method based on Convolutional Neural Networks (EDoF-CNN) that is able to rapidly perform the extended depth-of-field while also enhancing the spatial resolution of the resulting fused image. We evaluated our approach using simulated low-resolution z-stacks from Giemsa-stained thick blood smears from patients presenting with Plasmodium falciparum malaria. The EDoF-CNN allows speed-up of our digital-pathology acquisition platform and significantly improves the quality of the EDoF compared to the traditional multi-scaled approaches when applied to lower resolution stacks corresponding to acquisitions with fewer focal planes, large camera pixel binning or lower magnification objectives (larger FOV). We use the parasite detection accuracy of a deep learning model on the EDoFs as a concrete, task-specific measure of performance of this approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge