Amarda Shehu
Can Consumer Chatbots Reason? A Student-Led Field Experiment Embedded in an "AI-for-All" Undergraduate Course
Dec 28, 2025Abstract:Claims about whether large language model (LLM) chatbots "reason" are typically debated using curated benchmarks and laboratory-style evaluation protocols. This paper offers a complementary perspective: a student-led field experiment embedded as a midterm project in UNIV 182 (AI4All) at George Mason University, a Mason Core course designed for undergraduates across disciplines with no expected prior STEM exposure. Student teams designed their own reasoning tasks, ran them on widely used consumer chatbots representative of current capabilities, and evaluated both (i) answer correctness and (ii) the validity of the chatbot's stated reasoning (for example, cases where an answer is correct but the explanation is not, or vice versa). Across eight teams that reported standardized scores, students contributed 80 original reasoning prompts spanning six categories: pattern completion, transformation rules, spatial/visual reasoning, quantitative reasoning, relational/logic reasoning, and analogical reasoning. These prompts yielded 320 model responses plus follow-up explanations. Aggregating team-level results, OpenAI GPT-5 and Claude 4.5 achieved the highest mean answer accuracy (86.2% and 83.8%), followed by Grok 4 (82.5%) and Perplexity (73.1%); explanation validity showed a similar ordering (81.2%, 80.0%, 77.5%, 66.2%). Qualitatively, teams converged on a consistent error signature: strong performance on short, structured math and pattern items but reduced reliability on spatial/visual reasoning and multi-step transformations, with frequent "sound right but reason wrong" explanations. The assignment's primary contribution is pedagogical: it operationalizes AI literacy as experimental practice (prompt design, measurement, rater disagreement, and interpretability/grounding) while producing a reusable, student-generated corpus of reasoning probes grounded in authentic end-user interaction.
Foundation Models for AI-Enabled Biological Design
May 16, 2025Abstract:This paper surveys foundation models for AI-enabled biological design, focusing on recent developments in applying large-scale, self-supervised models to tasks such as protein engineering, small molecule design, and genomic sequence design. Though this domain is evolving rapidly, this survey presents and discusses a taxonomy of current models and methods. The focus is on challenges and solutions in adapting these models for biological applications, including biological sequence modeling architectures, controllability in generation, and multi-modal integration. The survey concludes with a discussion of open problems and future directions, offering concrete next-steps to improve the quality of biological sequence generation.
Birdie: Advancing State Space Models with Reward-Driven Objectives and Curricula
Nov 05, 2024



Abstract:Efficient state space models (SSMs), such as linear recurrent neural networks and linear attention variants, offer computational advantages over Transformers but struggle with tasks requiring long-range in-context retrieval-like text copying, associative recall, and question answering over long contexts. Previous efforts to address these challenges have focused on architectural modifications, often reintroducing computational inefficiencies. In this paper, we propose a novel training procedure, Birdie, that significantly enhances the in-context retrieval capabilities of SSMs without altering their architecture. Our approach combines bidirectional input processing with dynamic mixtures of specialized pre-training objectives, optimized via reinforcement learning. We introduce a new bidirectional SSM architecture that seamlessly transitions from bidirectional context processing to causal generation. Experimental evaluations demonstrate that Birdie markedly improves performance on retrieval-intensive tasks such as multi-number phone book lookup, long paragraph question-answering, and infilling. This narrows the performance gap with Transformers, while retaining computational efficiency. Our findings highlight the importance of training procedures in leveraging the fixed-state capacity of SSMs, offering a new direction to advance their capabilities. All code and pre-trained models are available at https://www.github.com/samblouir/birdie, with support for JAX and PyTorch.
Accounting for Work Zone Disruptions in Traffic Flow Forecasting
Jul 16, 2024Abstract:Traffic speed forecasting is an important task in intelligent transportation system management. The objective of much of the current computational research is to minimize the difference between predicted and actual speeds, but information modalities other than speed priors are largely not taken into account. In particular, though state of the art performance is achieved on speed forecasting with graph neural network methods, these methods do not incorporate information on roadway maintenance work zones and their impacts on predicted traffic flows; yet, the impacts of construction work zones are of significant interest to roadway management agencies, because they translate to impacts on the local economy and public well-being. In this paper, we build over the convolutional graph neural network architecture and present a novel ``Graph Convolutional Network for Roadway Work Zones" model that includes a novel data fusion mechanism and a new heterogeneous graph aggregation methodology to accommodate work zone information in spatio-temporal dependencies among traffic states. The model is evaluated on two data sets that capture traffic flows in the presence of work zones in the Commonwealth of Virginia. Extensive comparative evaluation and ablation studies show that the proposed model can capture complex and nonlinear spatio-temporal relationships across a transportation corridor, outperforming baseline models, particularly when predicting traffic flow during a workzone event.
Beyond Single-Model Views for Deep Learning: Optimization versus Generalizability of Stochastic Optimization Algorithms
Mar 01, 2024



Abstract:Despite an extensive body of literature on deep learning optimization, our current understanding of what makes an optimization algorithm effective is fragmented. In particular, we do not understand well whether enhanced optimization translates to improved generalizability. Current research overlooks the inherent stochastic nature of stochastic gradient descent (SGD) and its variants, resulting in a lack of comprehensive benchmarking and insight into their statistical performance. This paper aims to address this gap by adopting a novel approach. Rather than solely evaluating the endpoint of individual optimization trajectories, we draw from an ensemble of trajectories to estimate the stationary distribution of stochastic optimizers. Our investigation encompasses a wide array of techniques, including SGD and its variants, flat-minima optimizers, and new algorithms we propose under the Basin Hopping framework. Through our evaluation, which encompasses synthetic functions with known minima and real-world problems in computer vision and natural language processing, we emphasize fair benchmarking under a statistical framework, comparing stationary distributions and establishing statistical significance. Our study uncovers several key findings regarding the relationship between training loss and hold-out accuracy, as well as the comparable performance of SGD, noise-enabled variants, and novel optimizers utilizing the BH framework. Notably, these algorithms demonstrate performance on par with flat-minima optimizers like SAM, albeit with half the gradient evaluations. We anticipate that our work will catalyze further exploration in deep learning optimization, encouraging a shift away from single-model approaches towards methodologies that acknowledge and leverage the stochastic nature of optimizers.
Multi-objective Deep Data Generation with Correlated Property Control
Oct 06, 2022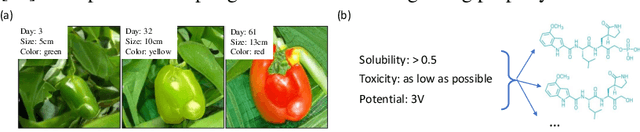
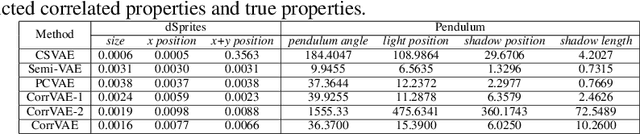
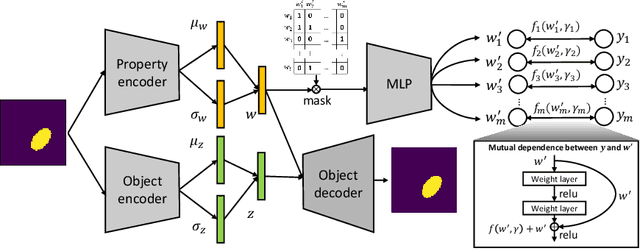
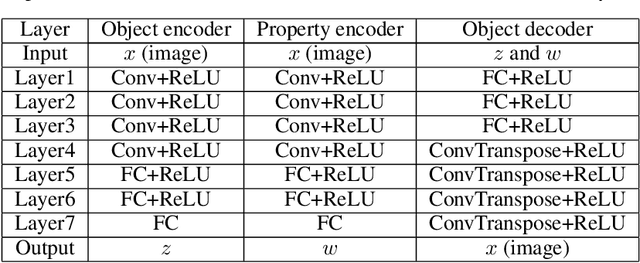
Abstract:Developing deep generative models has been an emerging field due to the ability to model and generate complex data for various purposes, such as image synthesis and molecular design. However, the advancement of deep generative models is limited by challenges to generate objects that possess multiple desired properties: 1) the existence of complex correlation among real-world properties is common but hard to identify; 2) controlling individual property enforces an implicit partially control of its correlated properties, which is difficult to model; 3) controlling multiple properties under various manners simultaneously is hard and under-explored. We address these challenges by proposing a novel deep generative framework that recovers semantics and the correlation of properties through disentangled latent vectors. The correlation is handled via an explainable mask pooling layer, and properties are precisely retained by generated objects via the mutual dependence between latent vectors and properties. Our generative model preserves properties of interest while handling correlation and conflicts of properties under a multi-objective optimization framework. The experiments demonstrate our model's superior performance in generating data with desired properties.
Multiple Instance Learning for Detecting Anomalies over Sequential Real-World Datasets
Oct 04, 2022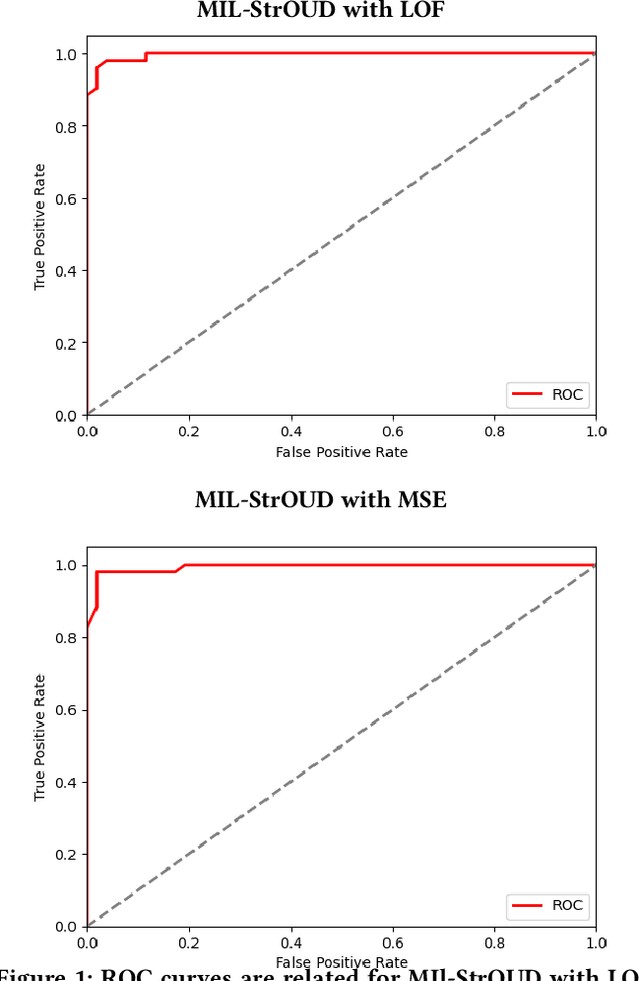
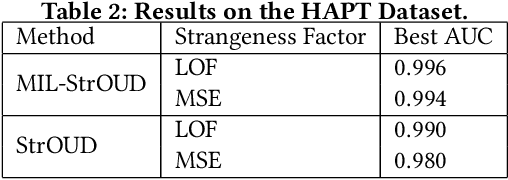
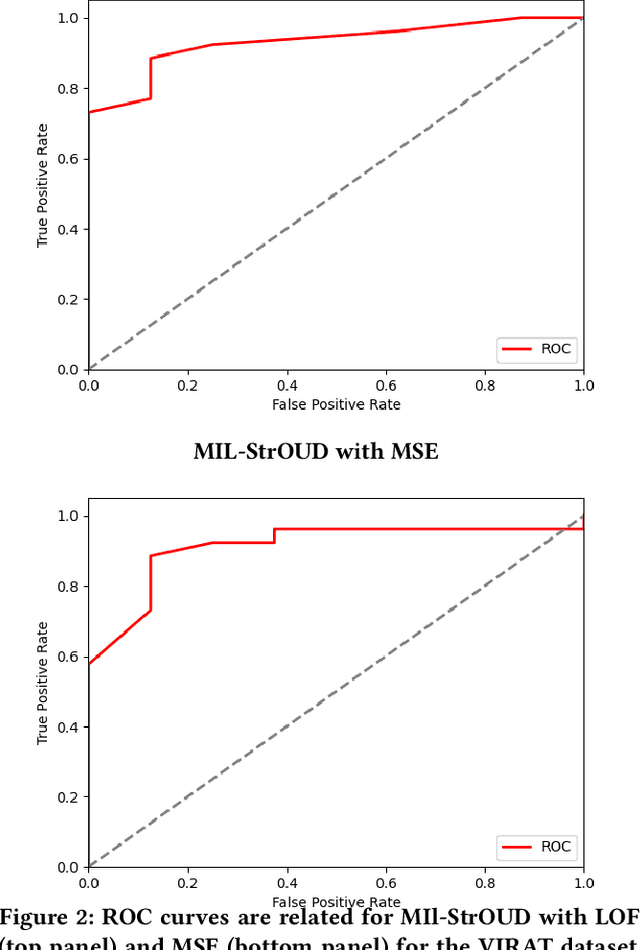
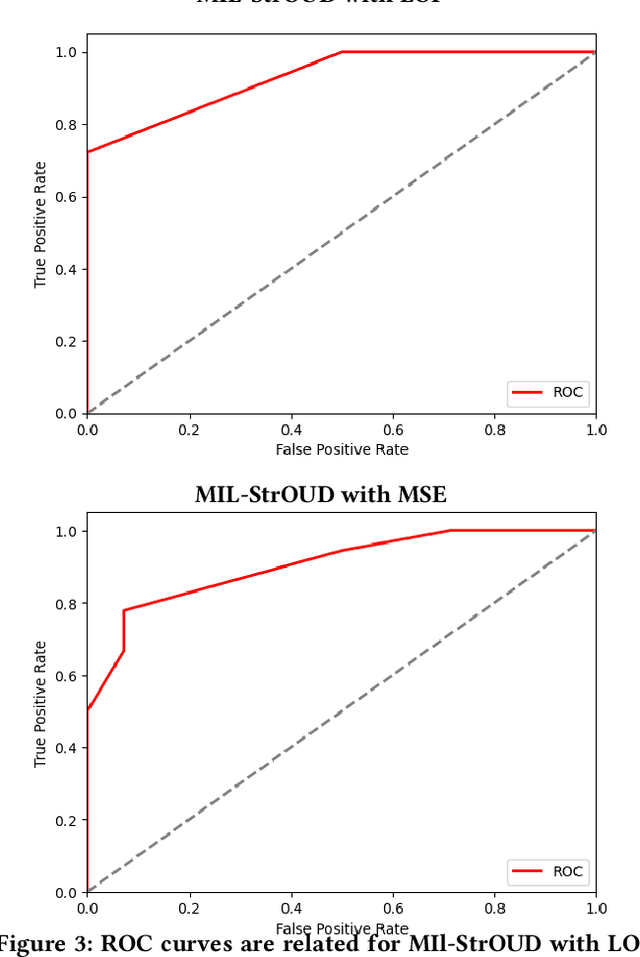
Abstract:Detecting anomalies over real-world datasets remains a challenging task. Data annotation is an intensive human labor problem, particularly in sequential datasets, where the start and end time of anomalies are not known. As a result, data collected from sequential real-world processes can be largely unlabeled or contain inaccurate labels. These characteristics challenge the application of anomaly detection techniques based on supervised learning. In contrast, Multiple Instance Learning (MIL) has been shown effective on problems with incomplete knowledge of labels in the training dataset, mainly due to the notion of bags. While largely under-leveraged for anomaly detection, MIL provides an appealing formulation for anomaly detection over real-world datasets, and it is the primary contribution of this paper. In this paper, we propose an MIL-based formulation and various algorithmic instantiations of this framework based on different design decisions for key components of the framework. We evaluate the resulting algorithms over four datasets that capture different physical processes along different modalities. The experimental evaluation draws out several observations. The MIL-based formulation performs no worse than single instance learning on easy to moderate datasets and outperforms single-instance learning on more challenging datasets. Altogether, the results show that the framework generalizes well over diverse datasets resulting from different real-world application domains.
Transformer Neural Networks Attending to Both Sequence and Structure for Protein Prediction Tasks
Jun 17, 2022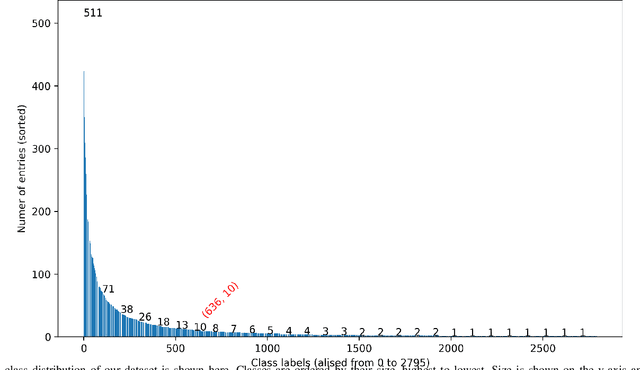
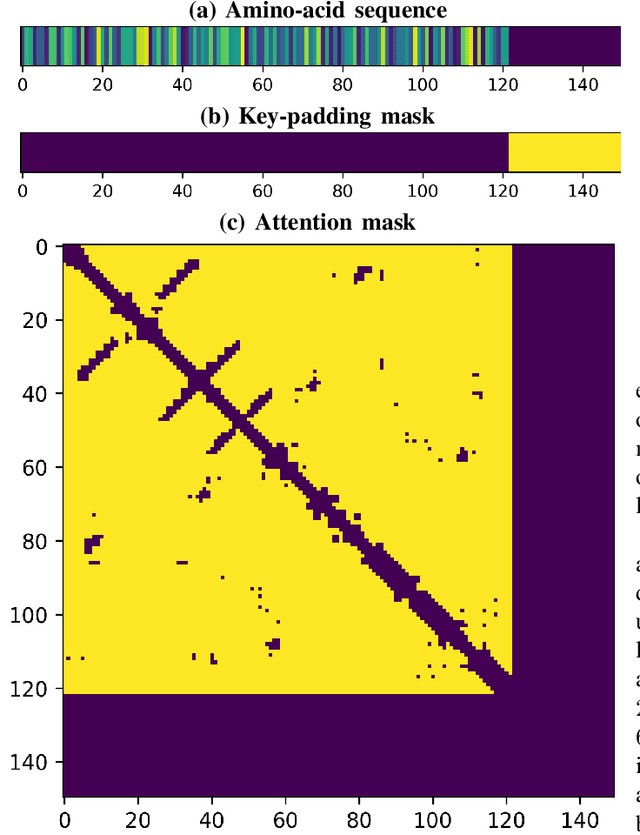
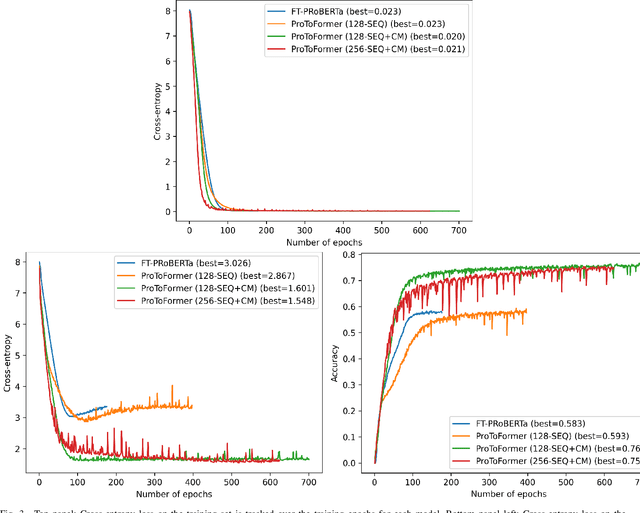
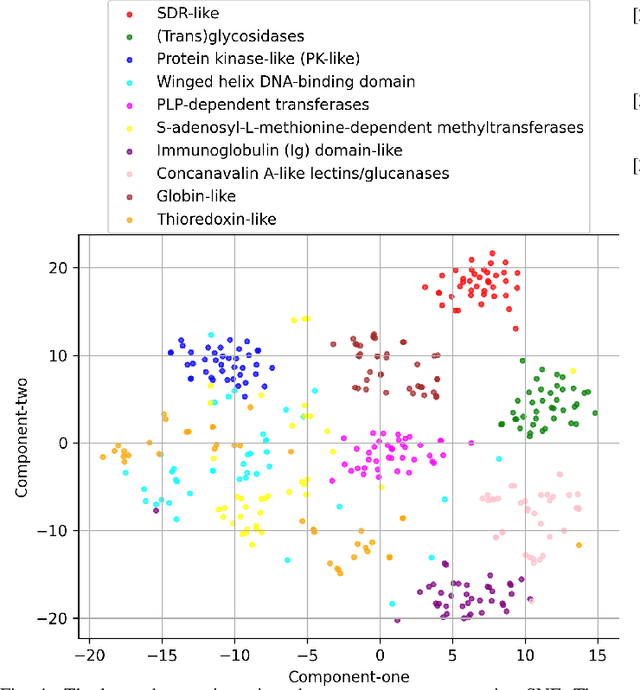
Abstract:The increasing number of protein sequences decoded from genomes is opening up new avenues of research on linking protein sequence to function with transformer neural networks. Recent research has shown that the number of known protein sequences supports learning useful, task-agnostic sequence representations via transformers. In this paper, we posit that learning joint sequence-structure representations yields better representations for function-related prediction tasks. We propose a transformer neural network that attends to both sequence and tertiary structure. We show that such joint representations are more powerful than sequence-based representations only, and they yield better performance on superfamily membership across various metrics.
Interpretable Molecular Graph Generation via Monotonic Constraints
Feb 28, 2022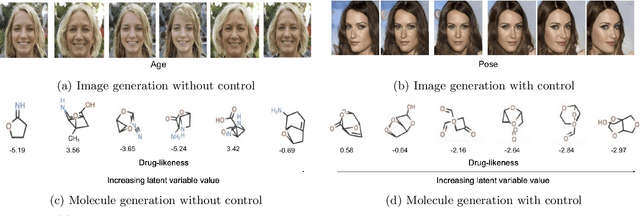
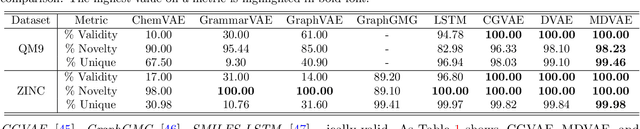
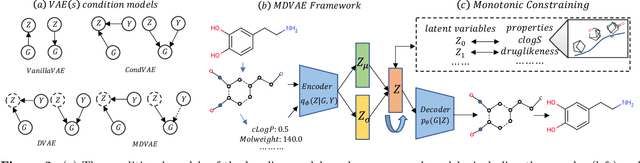
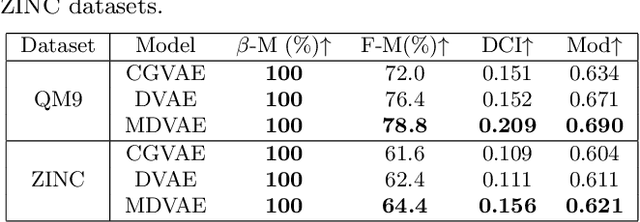
Abstract:Designing molecules with specific properties is a long-lasting research problem and is central to advancing crucial domains such as drug discovery and material science. Recent advances in deep graph generative models treat molecule design as graph generation problems which provide new opportunities toward the breakthrough of this long-lasting problem. Existing models, however, have many shortcomings, including poor interpretability and controllability toward desired molecular properties. This paper focuses on new methodologies for molecule generation with interpretable and controllable deep generative models, by proposing new monotonically-regularized graph variational autoencoders. The proposed models learn to represent the molecules with latent variables and then learn the correspondence between them and molecule properties parameterized by polynomial functions. To further improve the intepretability and controllability of molecule generation towards desired properties, we derive new objectives which further enforce monotonicity of the relation between some latent variables and target molecule properties such as toxicity and clogP. Extensive experimental evaluation demonstrates the superiority of the proposed framework on accuracy, novelty, disentanglement, and control towards desired molecular properties. The code is open-source at https://anonymous.4open.science/r/MDVAE-FD2C.
Traffic Flow Forecasting with Maintenance Downtime via Multi-Channel Attention-Based Spatio-Temporal Graph Convolutional Networks
Oct 04, 2021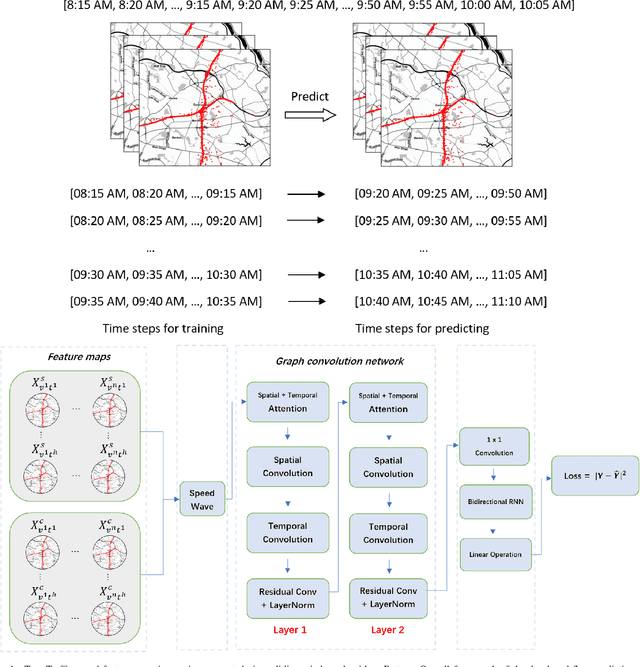
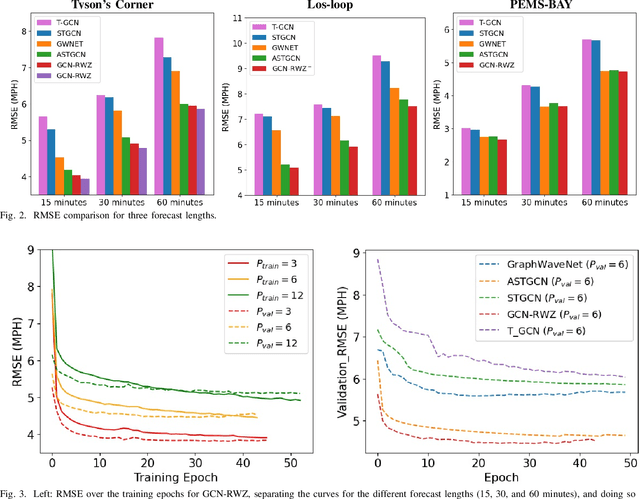
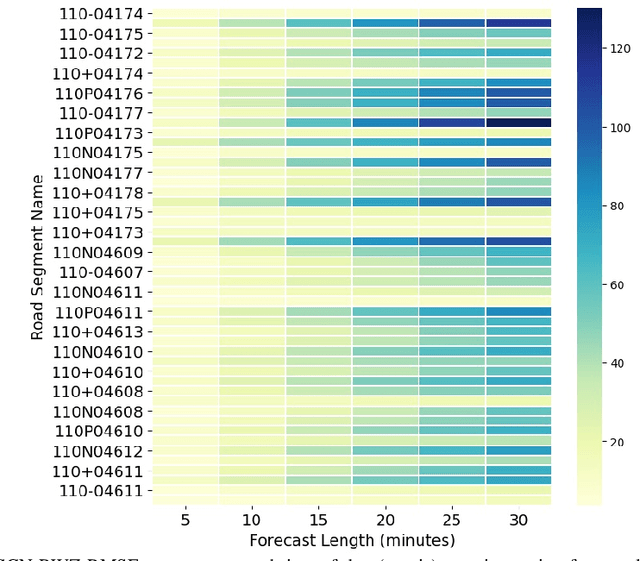
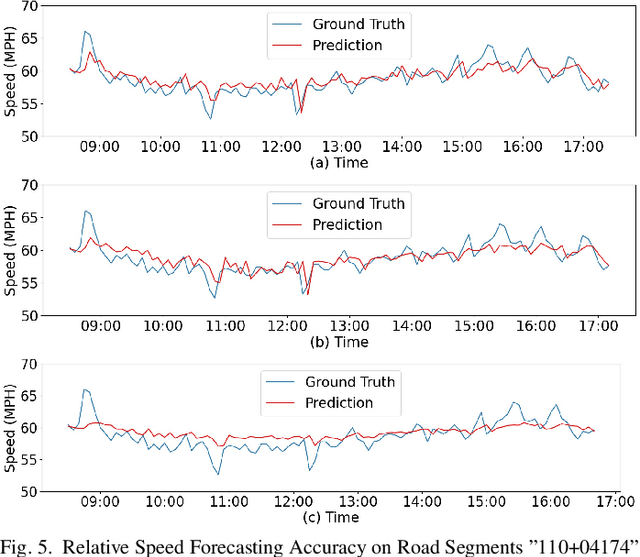
Abstract:Forecasting traffic flows is a central task in intelligent transportation system management. Graph structures have shown promise as a modeling framework, with recent advances in spatio-temporal modeling via graph convolution neural networks, improving the performance or extending the prediction horizon on traffic flows. However, a key shortcoming of state-of-the-art methods is their inability to take into account information of various modalities, for instance the impact of maintenance downtime on traffic flows. This is the issue we address in this paper. Specifically, we propose a novel model to predict traffic speed under the impact of construction work. The model is based on the powerful attention-based spatio-temporal graph convolution architecture but utilizes various channels to integrate different sources of information, explicitly builds spatio-temporal dependencies among traffic states, captures the relationships between heterogeneous roadway networks, and then predicts changes in traffic flow resulting from maintenance downtime events. The model is evaluated on two benchmark datasets and a novel dataset we have collected over the bustling Tyson's corner region in Northern Virginia. Extensive comparative experiments and ablation studies show that the proposed model can capture complex and nonlinear spatio-temporal relationships across a transportation corridor, outperforming baseline models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge