Ali Nasseri
BFS-Net: Weakly Supervised Cell Instance Segmentation from Bright-Field Microscopy Z-Stacks
Jun 09, 2022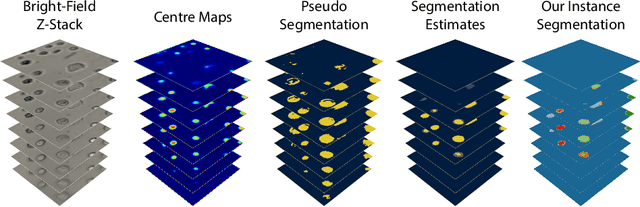
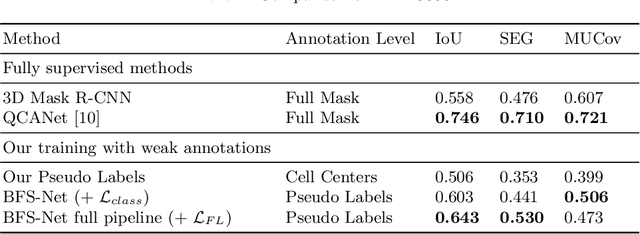
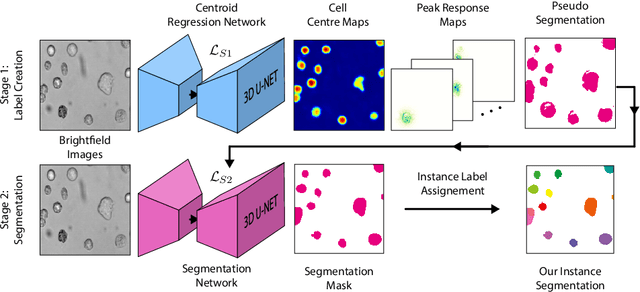
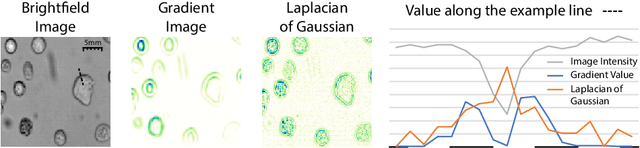
Abstract:Despite its broad availability, volumetric information acquisition from Bright-Field Microscopy (BFM) is inherently difficult due to the projective nature of the acquisition process. We investigate the prediction of 3D cell instances from a set of BFM Z-Stack images. We propose a novel two-stage weakly supervised method for volumetric instance segmentation of cells which only requires approximate cell centroids annotation. Created pseudo-labels are thereby refined with a novel refinement loss with Z-stack guidance. The evaluations show that our approach can generalize not only to BFM Z-Stack data, but to other 3D cell imaging modalities. A comparison of our pipeline against fully supervised methods indicates that the significant gain in reduced data collection and labelling results in minor performance difference.
Planning as Inference in Epidemiological Models
Apr 03, 2020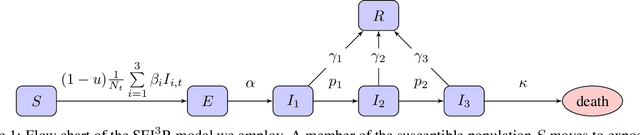
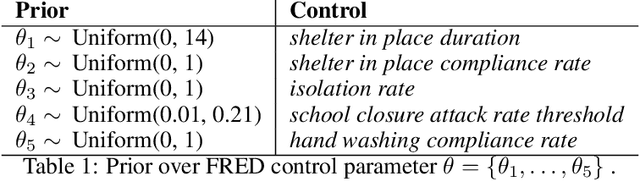
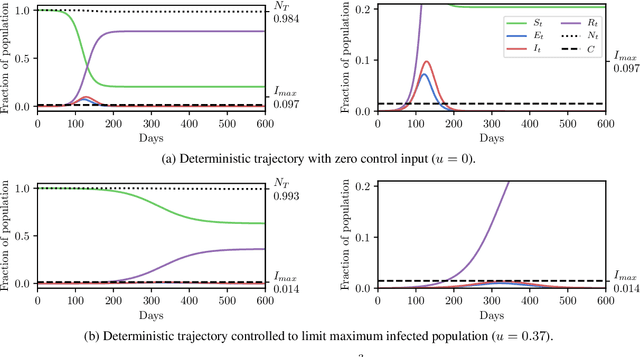
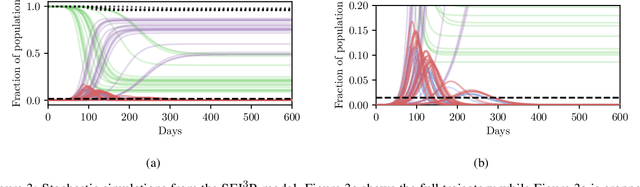
Abstract:In this work we demonstrate how existing software tools can be used to automate parts of infectious disease-control policy-making via performing inference in existing epidemiological dynamics models. The kind of inference tasks undertaken include computing, for planning purposes, the posterior distribution over putatively controllable, via direct policy-making choices, simulation model parameters that give rise to acceptable disease progression outcomes. Neither the full capabilities of such inference automation software tools nor their utility for planning is widely disseminated at the current time. Timely gains in understanding about these tools and how they can be used may lead to more fine-grained and less economically damaging policy prescriptions, particularly during the current COVID-19 pandemic.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge