Alaa Bessadok
Deep Cross-Modality and Resolution Graph Integration for Universal Brain Connectivity Mapping and Augmentation
Sep 13, 2022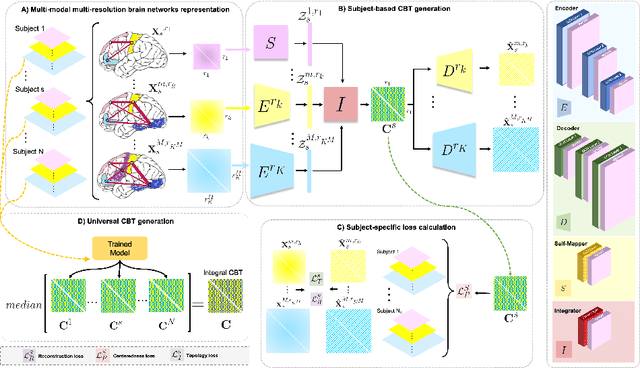
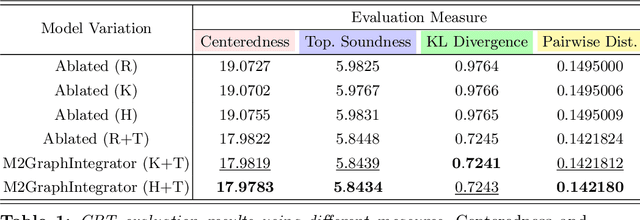
Abstract:The connectional brain template (CBT) captures the shared traits across all individuals of a given population of brain connectomes, thereby acting as a fingerprint. Estimating a CBT from a population where brain graphs are derived from diverse neuroimaging modalities (e.g., functional and structural) and at different resolutions (i.e., number of nodes) remains a formidable challenge to solve. Such network integration task allows for learning a rich and universal representation of the brain connectivity across varying modalities and resolutions. The resulting CBT can be substantially used to generate entirely new multimodal brain connectomes, which can boost the learning of the downs-stream tasks such as brain state classification. Here, we propose the Multimodal Multiresolution Brain Graph Integrator Network (i.e., M2GraphIntegrator), the first multimodal multiresolution graph integration framework that maps a given connectomic population into a well centered CBT. M2GraphIntegrator first unifies brain graph resolutions by utilizing resolution-specific graph autoencoders. Next, it integrates the resulting fixed-size brain graphs into a universal CBT lying at the center of its population. To preserve the population diversity, we further design a novel clustering-based training sample selection strategy which leverages the most heterogeneous training samples. To ensure the biological soundness of the learned CBT, we propose a topological loss that minimizes the topological gap between the ground-truth brain graphs and the learned CBT. Our experiments show that from a single CBT, one can generate realistic connectomic datasets including brain graphs of varying resolutions and modalities. We further demonstrate that our framework significantly outperforms benchmarks in reconstruction quality, augmentation task, centeredness and topological soundness.
Inter-Domain Alignment for Predicting High-Resolution Brain Networks Using Teacher-Student Learning
Oct 06, 2021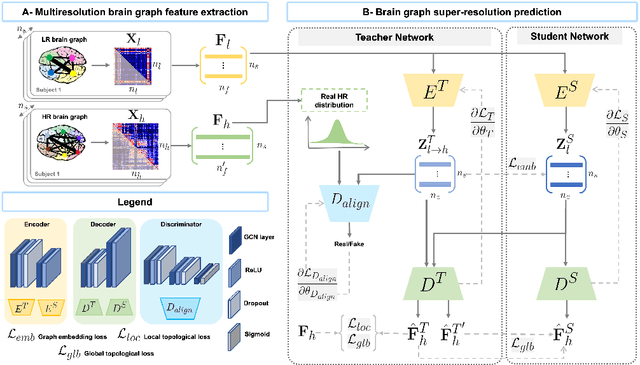
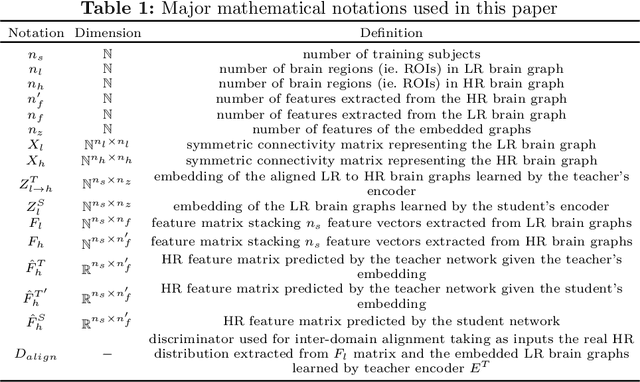
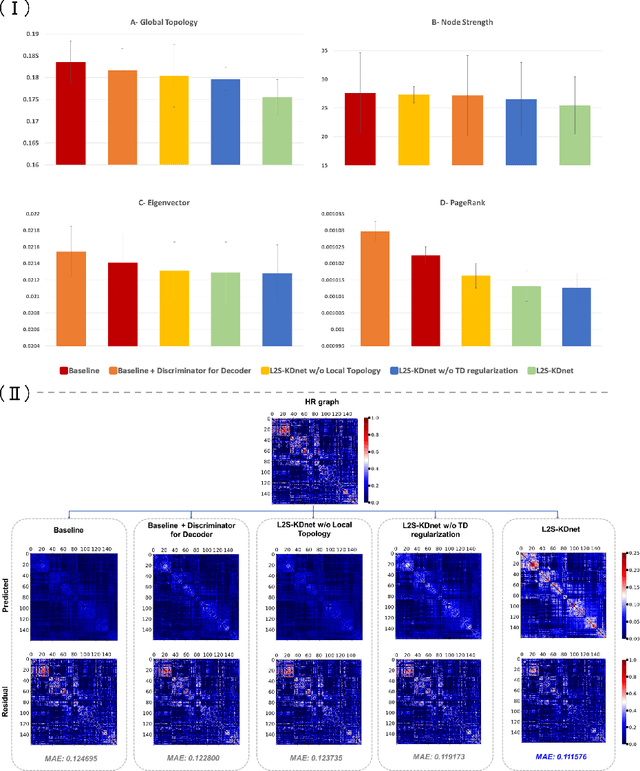
Abstract:Accurate and automated super-resolution image synthesis is highly desired since it has the great potential to circumvent the need for acquiring high-cost medical scans and a time-consuming preprocessing pipeline of neuroimaging data. However, existing deep learning frameworks are solely designed to predict high-resolution (HR) image from a low-resolution (LR) one, which limits their generalization ability to brain graphs (i.e., connectomes). A small body of works has focused on superresolving brain graphs where the goal is to predict a HR graph from a single LR graph. Although promising, existing works mainly focus on superresolving graphs belonging to the same domain (e.g., functional), overlooking the domain fracture existing between multimodal brain data distributions (e.g., morphological and structural). To this aim, we propose a novel inter-domain adaptation framework namely, Learn to SuperResolve Brain Graphs with Knowledge Distillation Network (L2S-KDnet), which adopts a teacher-student paradigm to superresolve brain graphs. Our teacher network is a graph encoder-decoder that firstly learns the LR brain graph embeddings, and secondly learns how to align the resulting latent representations to the HR ground truth data distribution using an adversarial regularization. Ultimately, it decodes the HR graphs from the aligned embeddings. Next, our student network learns the knowledge of the aligned brain graphs as well as the topological structure of the predicted HR graphs transferred from the teacher. We further leverage the decoder of the teacher to optimize the student network. L2S-KDnet presents the first TS architecture tailored for brain graph super-resolution synthesis that is based on inter-domain alignment. Our experimental results demonstrate substantial performance gains over benchmark methods.
A Few-shot Learning Graph Multi-Trajectory Evolution Network for Forecasting Multimodal Baby Connectivity Development from a Baseline Timepoint
Oct 06, 2021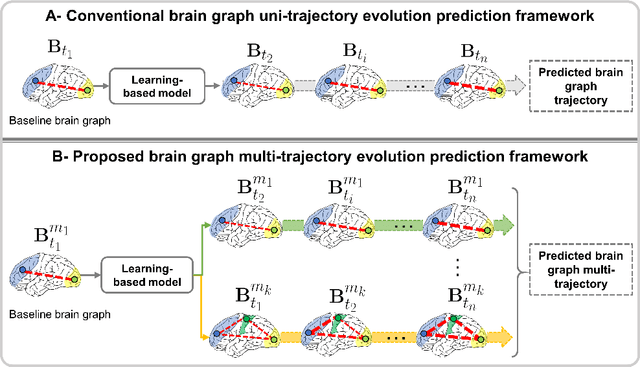
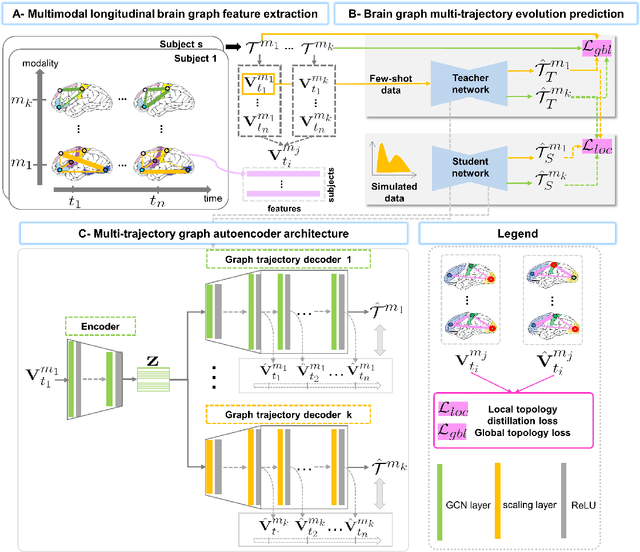
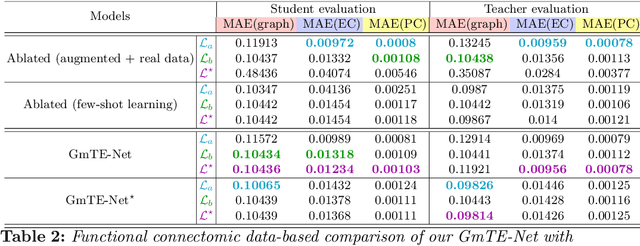
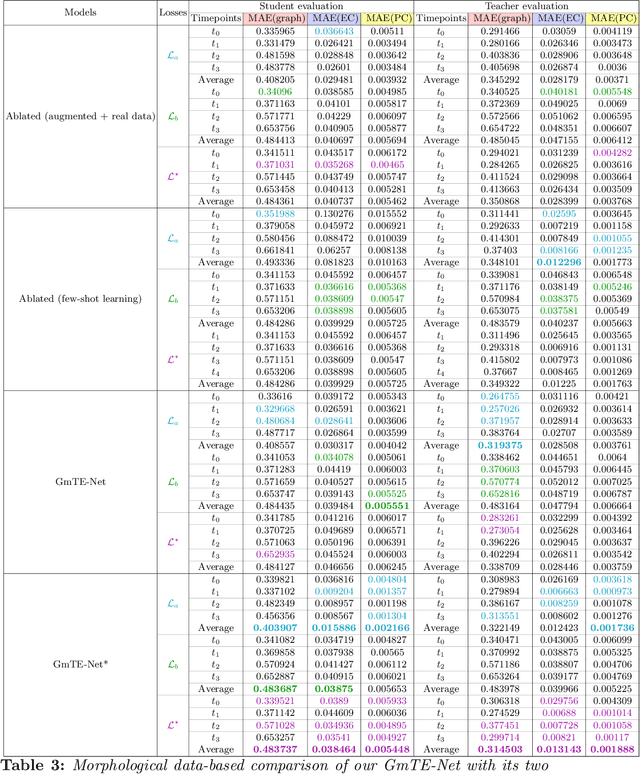
Abstract:Charting the baby connectome evolution trajectory during the first year after birth plays a vital role in understanding dynamic connectivity development of baby brains. Such analysis requires acquisition of longitudinal connectomic datasets. However, both neonatal and postnatal scans are rarely acquired due to various difficulties. A small body of works has focused on predicting baby brain evolution trajectory from a neonatal brain connectome derived from a single modality. Although promising, large training datasets are essential to boost model learning and to generalize to a multi-trajectory prediction from different modalities (i.e., functional and morphological connectomes). Here, we unprecedentedly explore the question: Can we design a few-shot learning-based framework for predicting brain graph trajectories across different modalities? To this aim, we propose a Graph Multi-Trajectory Evolution Network (GmTE-Net), which adopts a teacher-student paradigm where the teacher network learns on pure neonatal brain graphs and the student network learns on simulated brain graphs given a set of different timepoints. To the best of our knowledge, this is the first teacher-student architecture tailored for brain graph multi-trajectory growth prediction that is based on few-shot learning and generalized to graph neural networks (GNNs). To boost the performance of the student network, we introduce a local topology-aware distillation loss that forces the predicted graph topology of the student network to be consistent with the teacher network. Experimental results demonstrate substantial performance gains over benchmark methods. Hence, our GmTE-Net can be leveraged to predict atypical brain connectivity trajectory evolution across various modalities. Our code is available at https: //github.com/basiralab/GmTE-Net.
Graph Neural Networks in Network Neuroscience
Jun 07, 2021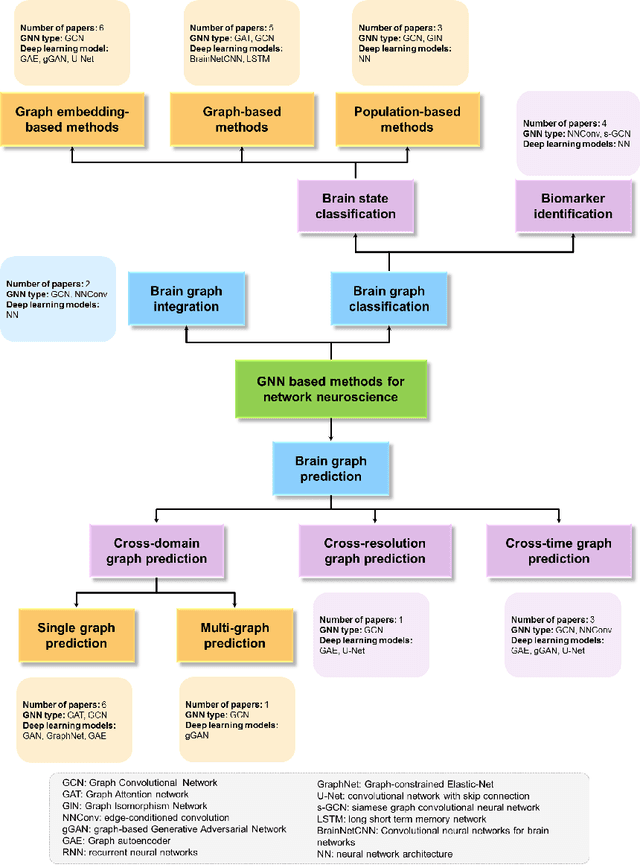
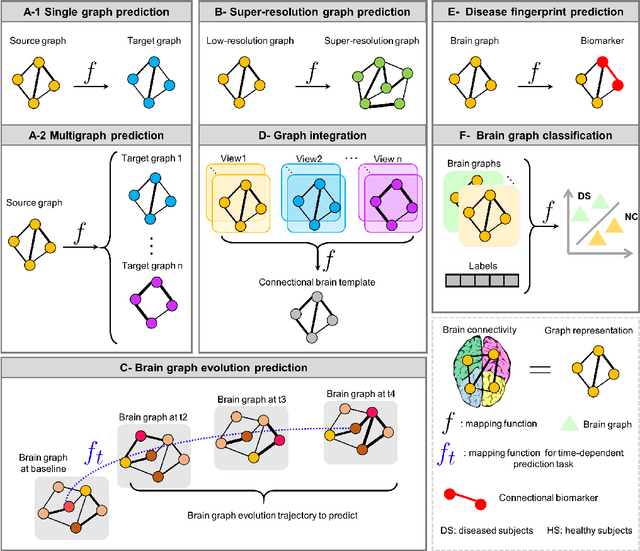
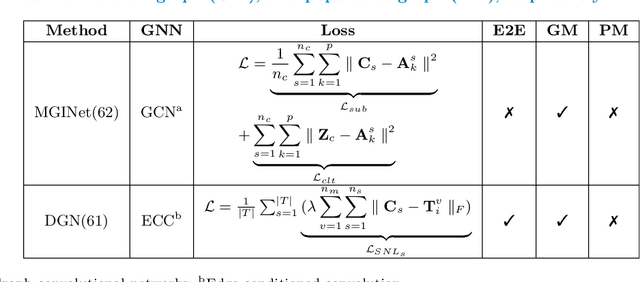
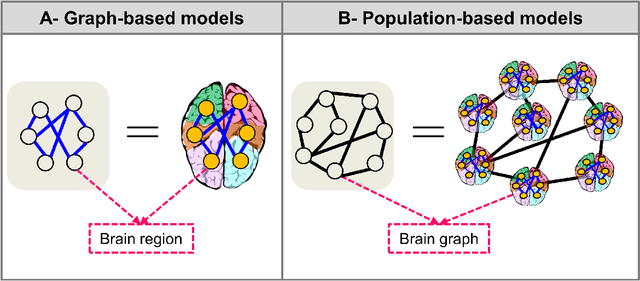
Abstract:Noninvasive medical neuroimaging has yielded many discoveries about the brain connectivity. Several substantial techniques mapping morphological, structural and functional brain connectivities were developed to create a comprehensive road map of neuronal activities in the human brain -namely brain graph. Relying on its non-Euclidean data type, graph neural network (GNN) provides a clever way of learning the deep graph structure and it is rapidly becoming the state-of-the-art leading to enhanced performance in various network neuroscience tasks. Here we review current GNN-based methods, highlighting the ways that they have been used in several applications related to brain graphs such as missing brain graph synthesis and disease classification. We conclude by charting a path toward a better application of GNN models in network neuroscience field for neurological disorder diagnosis and population graph integration. The list of papers cited in our work is available at https://github.com/basiralab/GNNs-in-Network-Neuroscience.
Brain Multigraph Prediction using Topology-Aware Adversarial Graph Neural Network
May 06, 2021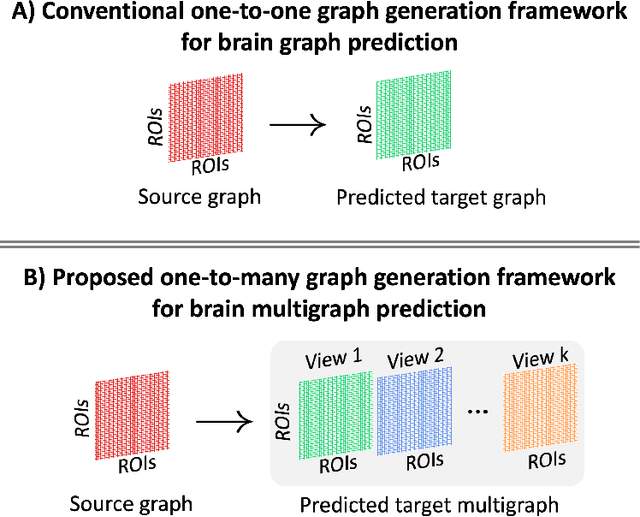
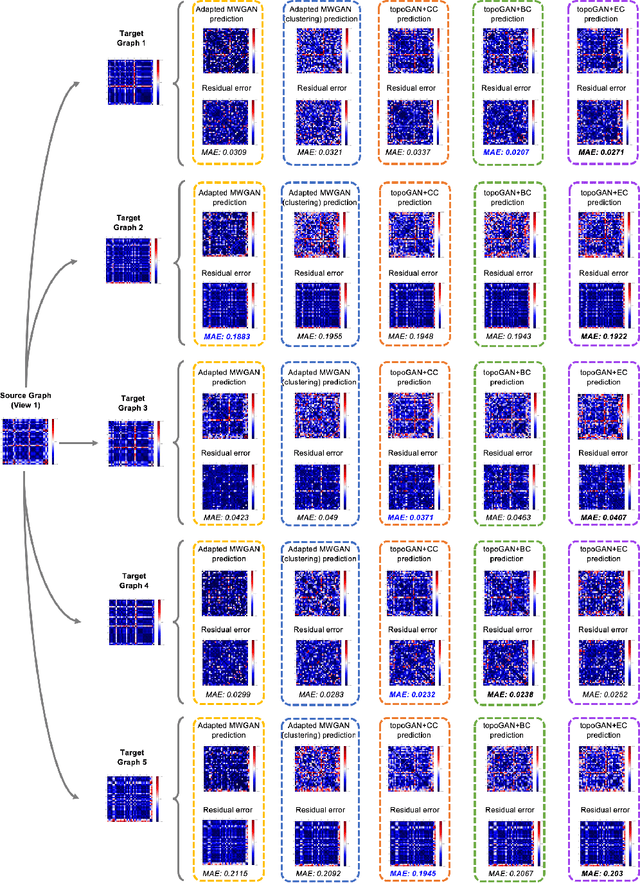
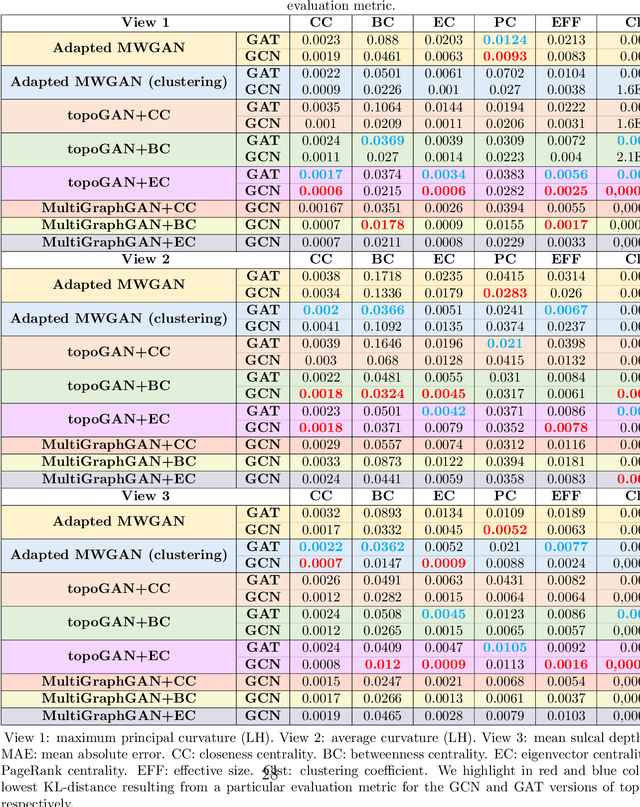
Abstract:Brain graphs (i.e, connectomes) constructed from medical scans such as magnetic resonance imaging (MRI) have become increasingly important tools to characterize the abnormal changes in the human brain. Due to the high acquisition cost and processing time of multimodal MRI, existing deep learning frameworks based on Generative Adversarial Network (GAN) focused on predicting the missing multimodal medical images from a few existing modalities. While brain graphs help better understand how a particular disorder can change the connectional facets of the brain, synthesizing a target brain multigraph (i.e, multiple brain graphs) from a single source brain graph is strikingly lacking. Additionally, existing graph generation works mainly learn one model for each target domain which limits their scalability in jointly predicting multiple target domains. Besides, while they consider the global topological scale of a graph (i.e., graph connectivity structure), they overlook the local topology at the node scale (e.g., how central a node is in the graph). To address these limitations, we introduce topology-aware graph GAN architecture (topoGAN), which jointly predicts multiple brain graphs from a single brain graph while preserving the topological structure of each target graph. Its three key innovations are: (i) designing a novel graph adversarial auto-encoder for predicting multiple brain graphs from a single one, (ii) clustering the encoded source graphs in order to handle the mode collapse issue of GAN and proposing a cluster-specific decoder, (iii) introducing a topological loss to force the prediction of topologically sound target brain graphs. The experimental results using five target domains demonstrated the outperformance of our method in brain multigraph prediction from a single graph in comparison with baseline approaches.
Residual Embedding Similarity-Based Network Selection for Predicting Brain Network Evolution Trajectory from a Single Observation
Sep 23, 2020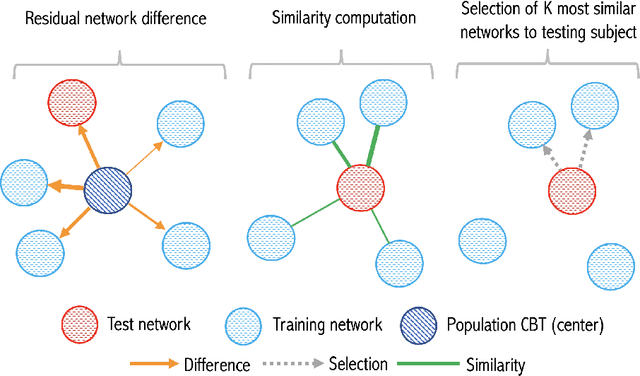
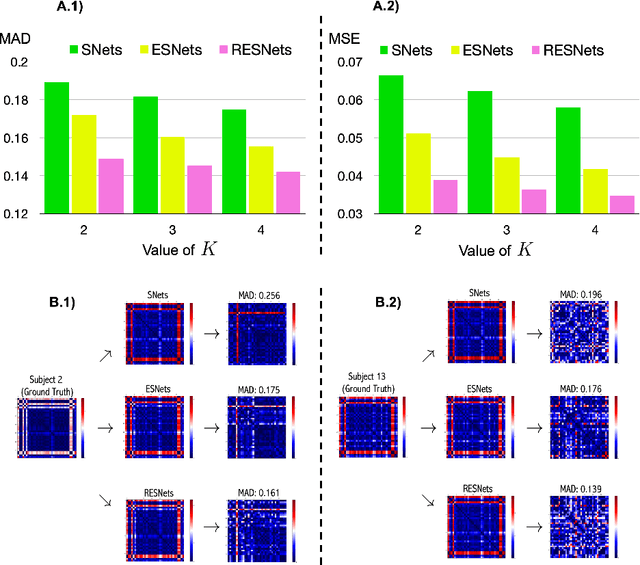
Abstract:While existing predictive frameworks are able to handle Euclidean structured data (i.e, brain images), they might fail to generalize to geometric non-Euclidean data such as brain networks. Besides, these are rooted the sample selection step in using Euclidean or learned similarity measure between vectorized training and testing brain networks. Such sample connectomic representation might include irrelevant and redundant features that could mislead the training sample selection step. Undoubtedly, this fails to exploit and preserve the topology of the brain connectome. To overcome this major drawback, we propose Residual Embedding Similarity-Based Network selection (RESNets) for predicting brain network evolution trajectory from a single timepoint. RESNets first learns a compact geometric embedding of each training and testing sample using adversarial connectome embedding network. This nicely reduces the high-dimensionality of brain networks while preserving their topological properties via graph convolutional networks. Next, to compute the similarity between subjects, we introduce the concept of a connectional brain template (CBT), a fixed network reference, where we further represent each training and testing network as a deviation from the reference CBT in the embedding space. As such, we select the most similar training subjects to the testing subject at baseline by comparing their learned residual embeddings with respect to the pre-defined CBT. Once the best training samples are selected at baseline, we simply average their corresponding brain networks at follow-up timepoints to predict the evolution trajectory of the testing network. Our experiments on both healthy and disordered brain networks demonstrate the success of our proposed method in comparison to RESNets ablated versions and traditional approaches.
Topology-Aware Generative Adversarial Network for Joint Prediction of Multiple Brain Graphs from a Single Brain Graph
Sep 23, 2020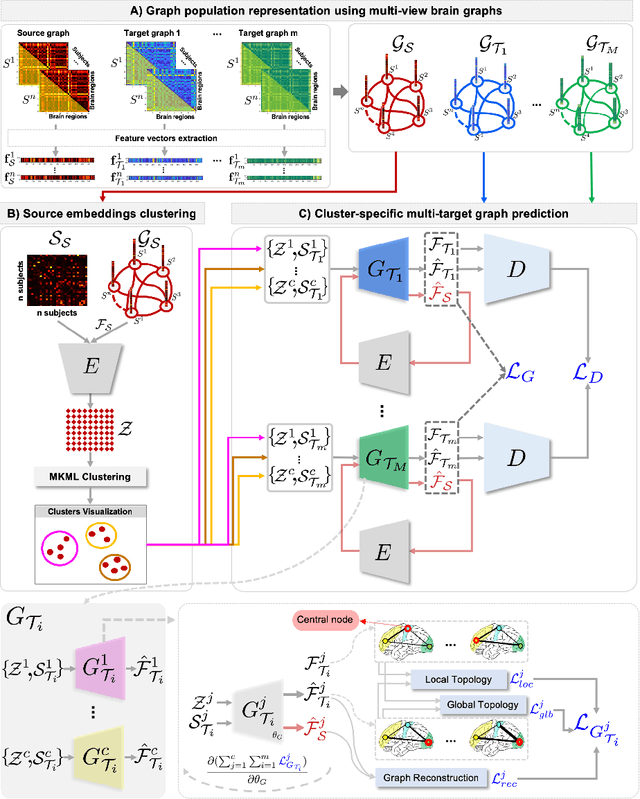

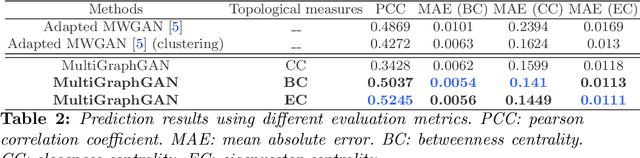
Abstract:Several works based on Generative Adversarial Networks (GAN) have been recently proposed to predict a set of medical images from a single modality (e.g, FLAIR MRI from T1 MRI). However, such frameworks are primarily designed to operate on images, limiting their generalizability to non-Euclidean geometric data such as brain graphs. While a growing number of connectomic studies has demonstrated the promise of including brain graphs for diagnosing neurological disorders, no geometric deep learning work was designed for multiple target brain graphs prediction from a source brain graph. Despite the momentum the field of graph generation has gained in the last two years, existing works have two critical drawbacks. First, the bulk of such works aims to learn one model for each target domain to generate from a source domain. Thus, they have a limited scalability in jointly predicting multiple target domains. Second, they merely consider the global topological scale of a graph (i.e., graph connectivity structure) and overlook the local topology at the node scale of a graph (e.g., how central a node is in the graph). To meet these challenges, we introduce MultiGraphGAN architecture, which not only predicts multiple brain graphs from a single brain graph but also preserves the topological structure of each target graph to predict. Its three core contributions lie in: (i) designing a graph adversarial auto-encoder for jointly predicting brain graphs from a single one, (ii) handling the mode collapse problem of GAN by clustering the encoded source graphs and proposing a cluster-specific decoder, (iii) introducing a topological loss to force the reconstruction of topologically sound target brain graphs. Our MultiGraphGAN significantly outperformed its variants thereby showing its great potential in multi-view brain graph generation from a single graph.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge