Chest Imagenome
Papers and Code
Anatomical grounding pre-training for medical phrase grounding
Feb 23, 2025Medical Phrase Grounding (MPG) maps radiological findings described in medical reports to specific regions in medical images. The primary obstacle hindering progress in MPG is the scarcity of annotated data available for training and validation. We propose anatomical grounding as an in-domain pre-training task that aligns anatomical terms with corresponding regions in medical images, leveraging large-scale datasets such as Chest ImaGenome. Our empirical evaluation on MS-CXR demonstrates that anatomical grounding pre-training significantly improves performance in both a zero-shot learning and fine-tuning setting, outperforming state-of-the-art MPG models. Our fine-tuned model achieved state-of-the-art performance on MS-CXR with an mIoU of 61.2, demonstrating the effectiveness of anatomical grounding pre-training for MPG.
Is Open-Source There Yet? A Comparative Study on Commercial and Open-Source LLMs in Their Ability to Label Chest X-Ray Reports
Feb 19, 2024



Introduction: With the rapid advances in large language models (LLMs), there have been numerous new open source as well as commercial models. While recent publications have explored GPT-4 in its application to extracting information of interest from radiology reports, there has not been a real-world comparison of GPT-4 to different leading open-source models. Materials and Methods: Two different and independent datasets were used. The first dataset consists of 540 chest x-ray reports that were created at the Massachusetts General Hospital between July 2019 and July 2021. The second dataset consists of 500 chest x-ray reports from the ImaGenome dataset. We then compared the commercial models GPT-3.5 Turbo and GPT-4 from OpenAI to the open-source models Mistral-7B, Mixtral-8x7B, Llama2-13B, Llama2-70B, QWEN1.5-72B and CheXbert and CheXpert-labeler in their ability to accurately label the presence of multiple findings in x-ray text reports using different prompting techniques. Results: On the ImaGenome dataset, the best performing open-source model was Llama2-70B with micro F1-scores of 0.972 and 0.970 for zero- and few-shot prompts, respectively. GPT-4 achieved micro F1-scores of 0.975 and 0.984, respectively. On the institutional dataset, the best performing open-source model was QWEN1.5-72B with micro F1-scores of 0.952 and 0.965 for zero- and few-shot prompting, respectively. GPT-4 achieved micro F1-scores of 0.975 and 0.973, respectively. Conclusion: In this paper, we show that while GPT-4 is superior to open-source models in zero-shot report labeling, the implementation of few-shot prompting can bring open-source models on par with GPT-4. This shows that open-source models could be a performant and privacy preserving alternative to GPT-4 for the task of radiology report classification.
Region-based Contrastive Pretraining for Medical Image Retrieval with Anatomic Query
May 09, 2023



We introduce a novel Region-based contrastive pretraining for Medical Image Retrieval (RegionMIR) that demonstrates the feasibility of medical image retrieval with similar anatomical regions. RegionMIR addresses two major challenges for medical image retrieval i) standardization of clinically relevant searching criteria (e.g., anatomical, pathology-based), and ii) localization of anatomical area of interests that are semantically meaningful. In this work, we propose an ROI image retrieval image network that retrieves images with similar anatomy by extracting anatomical features (via bounding boxes) and evaluate similarity between pairwise anatomy-categorized features between the query and the database of images using contrastive learning. ROI queries are encoded using a contrastive-pretrained encoder that was fine-tuned for anatomy classification, which generates an anatomical-specific latent space for region-correlated image retrieval. During retrieval, we compare the anatomically encoded query to find similar features within a feature database generated from training samples, and retrieve images with similar regions from training samples. We evaluate our approach on both anatomy classification and image retrieval tasks using the Chest ImaGenome Dataset. Our proposed strategy yields an improvement over state-of-the-art pretraining and co-training strategies, from 92.24 to 94.12 (2.03%) classification accuracy in anatomies. We qualitatively evaluate the image retrieval performance demonstrating generalizability across multiple anatomies with different morphology.
CheXRelNet: An Anatomy-Aware Model for Tracking Longitudinal Relationships between Chest X-Rays
Aug 08, 2022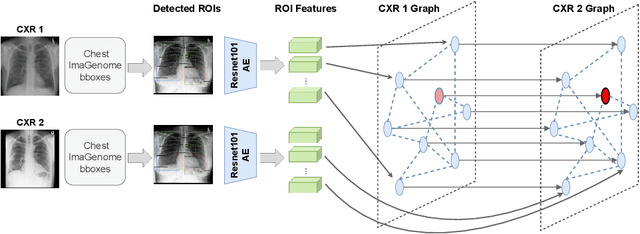
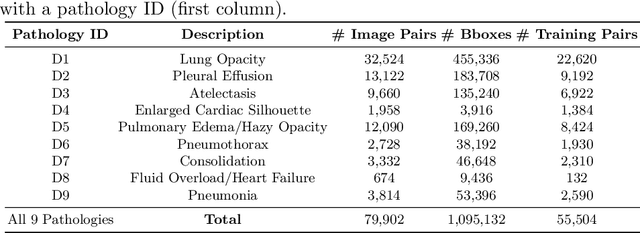
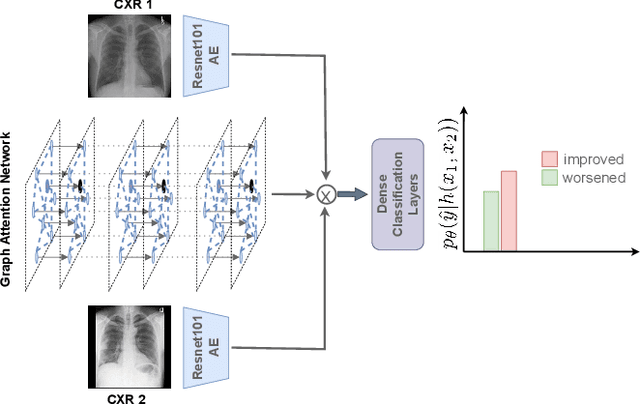
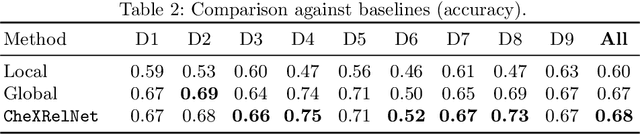
Despite the progress in utilizing deep learning to automate chest radiograph interpretation and disease diagnosis tasks, change between sequential Chest X-rays (CXRs) has received limited attention. Monitoring the progression of pathologies that are visualized through chest imaging poses several challenges in anatomical motion estimation and image registration, i.e., spatially aligning the two images and modeling temporal dynamics in change detection. In this work, we propose CheXRelNet, a neural model that can track longitudinal pathology change relations between two CXRs. CheXRelNet incorporates local and global visual features, utilizes inter-image and intra-image anatomical information, and learns dependencies between anatomical region attributes, to accurately predict disease change for a pair of CXRs. Experimental results on the Chest ImaGenome dataset show increased downstream performance compared to baselines. Code is available at https://github.com/PLAN-Lab/ChexRelNet
Chest ImaGenome Dataset for Clinical Reasoning
Jul 31, 2021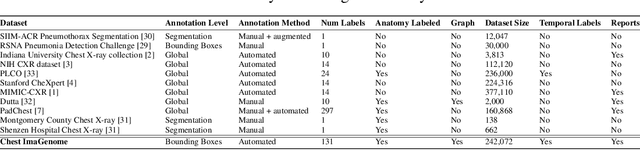
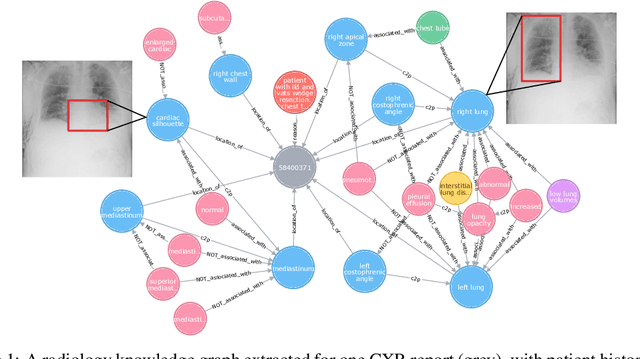
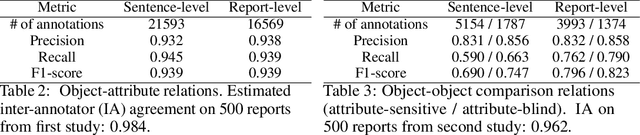
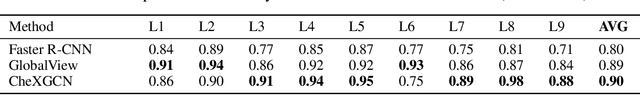
Despite the progress in automatic detection of radiologic findings from chest X-ray (CXR) images in recent years, a quantitative evaluation of the explainability of these models is hampered by the lack of locally labeled datasets for different findings. With the exception of a few expert-labeled small-scale datasets for specific findings, such as pneumonia and pneumothorax, most of the CXR deep learning models to date are trained on global "weak" labels extracted from text reports, or trained via a joint image and unstructured text learning strategy. Inspired by the Visual Genome effort in the computer vision community, we constructed the first Chest ImaGenome dataset with a scene graph data structure to describe $242,072$ images. Local annotations are automatically produced using a joint rule-based natural language processing (NLP) and atlas-based bounding box detection pipeline. Through a radiologist constructed CXR ontology, the annotations for each CXR are connected as an anatomy-centered scene graph, useful for image-level reasoning and multimodal fusion applications. Overall, we provide: i) $1,256$ combinations of relation annotations between $29$ CXR anatomical locations (objects with bounding box coordinates) and their attributes, structured as a scene graph per image, ii) over $670,000$ localized comparison relations (for improved, worsened, or no change) between the anatomical locations across sequential exams, as well as ii) a manually annotated gold standard scene graph dataset from $500$ unique patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge