Automatic Liver And Tumor Segmentation
Papers and Code
Precise Liver Tumor Segmentation in CT Using a Hybrid Deep Learning-Radiomics Framework
Dec 08, 2025Accurate three-dimensional delineation of liver tumors on contrast-enhanced CT is a prerequisite for treatment planning, navigation and response assessment, yet manual contouring is slow, observer-dependent and difficult to standardise across centres. Automatic segmentation is complicated by low lesion-parenchyma contrast, blurred or incomplete boundaries, heterogeneous enhancement patterns, and confounding structures such as vessels and adjacent organs. We propose a hybrid framework that couples an attention-enhanced cascaded U-Net with handcrafted radiomics and voxel-wise 3D CNN refinement for joint liver and liver-tumor segmentation. First, a 2.5D two-stage network with a densely connected encoder, sub-pixel convolution decoders and multi-scale attention gates produces initial liver and tumor probability maps from short stacks of axial slices. Inter-slice temporal consistency is then enforced by a simple three-slice refinement rule along the cranio-caudal direction, which restores thin and tiny lesions while suppressing isolated noise. Next, 728 radiomic descriptors spanning intensity, texture, shape, boundary and wavelet feature groups are extracted from candidate lesions and reduced to 20 stable, highly informative features via multi-strategy feature selection; a random forest classifier uses these features to reject false-positive regions. Finally, a compact 3D patch-based CNN derived from AlexNet operates in a narrow band around the tumor boundary to perform voxel-level relabelling and contour smoothing.
Navigated hepatic tumor resection using intraoperative ultrasound imaging
Oct 31, 2025Purpose: This proof-of-concept study evaluates feasibility and accuracy of an ultrasound-based navigation system for open liver surgery. Unlike most conventional systems that rely on registration to preoperative imaging, the proposed system provides navigation-guided resection using 3D models generated from intraoperative ultrasound. Methods: A pilot study was conducted in 25 patients undergoing resection of liver metastases. The first five cases served to optimize the workflow. Intraoperatively, an electromagnetic sensor compensated for organ motion, after which an ultrasound volume was acquired. Vasculature was segmented automatically and tumors semi-automatically using region-growing (n=15) or a deep learning algorithm (n=5). The resulting 3D model was visualized alongside tracked surgical instruments. Accuracy was assessed by comparing the distance between surgical clips and tumors in the navigation software with the same distance on a postoperative CT of the resected specimen. Results: Navigation was successfully established in all 20 patients. However, four cases were excluded from accuracy assessment due to intraoperative sensor detachment (n=3) or incorrect data recording (n=1). The complete navigation workflow was operational within 5-10 minutes. In 16 evaluable patients, 78 clip-to-tumor distances were analyzed. The median navigation accuracy was 3.2 mm [IQR: 2.8-4.8 mm], and an R0 resection was achieved in 15/16 (93.8%) patients and one patient had an R1 vascular resection. Conclusion: Navigation based solely on intra-operative ultrasound is feasible and accurate for liver surgery. This registration-free approach paves the way for simpler and more accurate image guidance systems.
Automatic segmentation of colorectal liver metastases for ultrasound-based navigated resection
Nov 07, 2025



Introduction: Accurate intraoperative delineation of colorectal liver metastases (CRLM) is crucial for achieving negative resection margins but remains challenging using intraoperative ultrasound (iUS) due to low contrast, noise, and operator dependency. Automated segmentation could enhance precision and efficiency in ultrasound-based navigation workflows. Methods: Eighty-five tracked 3D iUS volumes from 85 CRLM patients were used to train and evaluate a 3D U-Net implemented via the nnU-Net framework. Two variants were compared: one trained on full iUS volumes and another on cropped regions around tumors. Segmentation accuracy was assessed using Dice Similarity Coefficient (DSC), Hausdorff Distance (HDist.), and Relative Volume Difference (RVD) on retrospective and prospective datasets. The workflow was integrated into 3D Slicer for real-time intraoperative use. Results: The cropped-volume model significantly outperformed the full-volume model across all metrics (AUC-ROC = 0.898 vs 0.718). It achieved median DSC = 0.74, recall = 0.79, and HDist. = 17.1 mm comparable to semi-automatic segmentation but with ~4x faster execution (~ 1 min). Prospective intraoperative testing confirmed robust and consistent performance, with clinically acceptable accuracy for real-time surgical guidance. Conclusion: Automatic 3D segmentation of CRLM in iUS using a cropped 3D U-Net provides reliable, near real-time results with minimal operator input. The method enables efficient, registration-free ultrasound-based navigation for hepatic surgery, approaching expert-level accuracy while substantially reducing manual workload and procedure time.
Deep Learning-Based Automatic Delineation of Liver Domes in kV Triggered Images for Online Breath-hold Reproducibility Verification of Liver Stereotactic Body Radiation Therapy
Nov 22, 2024



Stereotactic Body Radiation Therapy (SBRT) can be a precise, minimally invasive treatment method for liver cancer and liver metastases. However, the effectiveness of SBRT relies on the accurate delivery of the dose to the tumor while sparing healthy tissue. Challenges persist in ensuring breath-hold reproducibility, with current methods often requiring manual verification of liver dome positions from kV-triggered images. To address this, we propose a proof-of-principle study of a deep learning-based pipeline to automatically delineate the liver dome from kV-planar images. From 24 patients who received SBRT for liver cancer or metastasis inside liver, 711 KV-triggered images acquired for online breath-hold verification were included in the current study. We developed a pipeline comprising a trained U-Net for automatic liver dome region segmentation from the triggered images followed by extraction of the liver dome via thresholding, edge detection, and morphological operations. The performance and generalizability of the pipeline was evaluated using 2-fold cross validation. The training of the U-Net model for liver region segmentation took under 30 minutes and the automatic delineation of a liver dome for any triggered image took less than one second. The RMSE and rate of detection for Fold1 with 366 images was (6.4 +/- 1.6) mm and 91.7%, respectively. For Fold2 with 345 images, the RMSE and rate of detection was (7.7 +/- 2.3) mm and 76.3% respectively.
CAFCT: Contextual and Attentional Feature Fusions of Convolutional Neural Networks and Transformer for Liver Tumor Segmentation
Jan 30, 2024



Medical image semantic segmentation techniques can help identify tumors automatically from computed tomography (CT) scans. In this paper, we propose a Contextual and Attentional feature Fusions enhanced Convolutional Neural Network (CNN) and Transformer hybrid network (CAFCT) model for liver tumor segmentation. In the proposed model, three other modules are introduced in the network architecture: Attentional Feature Fusion (AFF), Atrous Spatial Pyramid Pooling (ASPP) of DeepLabv3, and Attention Gates (AGs) to improve contextual information related to tumor boundaries for accurate segmentation. Experimental results show that the proposed CAFCT achieves a mean Intersection over Union (IoU) of 90.38% and Dice score of 86.78%, respectively, on the Liver Tumor Segmentation Benchmark (LiTS) dataset, outperforming pure CNN or Transformer methods, e.g., Attention U-Net, and PVTFormer.
MFA-Net: Multi-Scale feature fusion attention network for liver tumor segmentation
May 07, 2024



Segmentation of organs of interest in medical CT images is beneficial for diagnosis of diseases. Though recent methods based on Fully Convolutional Neural Networks (F-CNNs) have shown success in many segmentation tasks, fusing features from images with different scales is still a challenge: (1) Due to the lack of spatial awareness, F-CNNs share the same weights at different spatial locations. (2) F-CNNs can only obtain surrounding information through local receptive fields. To address the above challenge, we propose a new segmentation framework based on attention mechanisms, named MFA-Net (Multi-Scale Feature Fusion Attention Network). The proposed framework can learn more meaningful feature maps among multiple scales and result in more accurate automatic segmentation. We compare our proposed MFA-Net with SOTA methods on two 2D liver CT datasets. The experimental results show that our MFA-Net produces more precise segmentation on images with different scales.
Anisotropic Hybrid Networks for liver tumor segmentation with uncertainty quantification
Aug 23, 2023



The burden of liver tumors is important, ranking as the fourth leading cause of cancer mortality. In case of hepatocellular carcinoma (HCC), the delineation of liver and tumor on contrast-enhanced magnetic resonance imaging (CE-MRI) is performed to guide the treatment strategy. As this task is time-consuming, needs high expertise and could be subject to inter-observer variability there is a strong need for automatic tools. However, challenges arise from the lack of available training data, as well as the high variability in terms of image resolution and MRI sequence. In this work we propose to compare two different pipelines based on anisotropic models to obtain the segmentation of the liver and tumors. The first pipeline corresponds to a baseline multi-class model that performs the simultaneous segmentation of the liver and tumor classes. In the second approach, we train two distinct binary models, one segmenting the liver only and the other the tumors. Our results show that both pipelines exhibit different strengths and weaknesses. Moreover we propose an uncertainty quantification strategy allowing the identification of potential false positive tumor lesions. Both solutions were submitted to the MICCAI 2023 Atlas challenge regarding liver and tumor segmentation.
Encoding feature supervised UNet++: Redesigning Supervision for liver and tumor segmentation
Nov 15, 2022Liver tumor segmentation in CT images is a critical step in the diagnosis, surgical planning and postoperative evaluation of liver disease. An automatic liver and tumor segmentation method can greatly relieve physicians of the heavy workload of examining CT images and better improve the accuracy of diagnosis. In the last few decades, many modifications based on U-Net model have been proposed in the literature. However, there are relatively few improvements for the advanced UNet++ model. In our paper, we propose an encoding feature supervised UNet++(ES-UNet++) and apply it to the liver and tumor segmentation. ES-UNet++ consists of an encoding UNet++ and a segmentation UNet++. The well-trained encoding UNet++ can extract the encoding features of label map which are used to additionally supervise the segmentation UNet++. By adding supervision to the each encoder of segmentation UNet++, U-Nets of different depths that constitute UNet++ outperform the original version by average 5.7% in dice score and the overall dice score is thus improved by 2.1%. ES-UNet++ is evaluated with dataset LiTS, achieving 95.6% for liver segmentation and 67.4% for tumor segmentation in dice score. In this paper, we also concluded some valuable properties of ES-UNet++ by conducting comparative anaylsis between ES-UNet++ and UNet++:(1) encoding feature supervision can accelerate the convergence of the model.(2) encoding feature supervision enhances the effect of model pruning by achieving huge speedup while providing pruned models with fairly good performance.
3D Medical Image Segmentation based on multi-scale MPU-Net
Jul 24, 2023



The high cure rate of cancer is inextricably linked to physicians' accuracy in diagnosis and treatment, therefore a model that can accomplish high-precision tumor segmentation has become a necessity in many applications of the medical industry. It can effectively lower the rate of misdiagnosis while considerably lessening the burden on clinicians. However, fully automated target organ segmentation is problematic due to the irregular stereo structure of 3D volume organs. As a basic model for this class of real applications, U-Net excels. It can learn certain global and local features, but still lacks the capacity to grasp spatial long-range relationships and contextual information at multiple scales. This paper proposes a tumor segmentation model MPU-Net for patient volume CT images, which is inspired by Transformer with a global attention mechanism. By combining image serialization with the Position Attention Module, the model attempts to comprehend deeper contextual dependencies and accomplish precise positioning. Each layer of the decoder is also equipped with a multi-scale module and a cross-attention mechanism. The capability of feature extraction and integration at different levels has been enhanced, and the hybrid loss function developed in this study can better exploit high-resolution characteristic information. Moreover, the suggested architecture is tested and evaluated on the Liver Tumor Segmentation Challenge 2017 (LiTS 2017) dataset. Compared with the benchmark model U-Net, MPU-Net shows excellent segmentation results. The dice, accuracy, precision, specificity, IOU, and MCC metrics for the best model segmentation results are 92.17%, 99.08%, 91.91%, 99.52%, 85.91%, and 91.74%, respectively. Outstanding indicators in various aspects illustrate the exceptional performance of this framework in automatic medical image segmentation.
Free-form Lesion Synthesis Using a Partial Convolution Generative Adversarial Network for Enhanced Deep Learning Liver Tumor Segmentation
Jun 18, 2022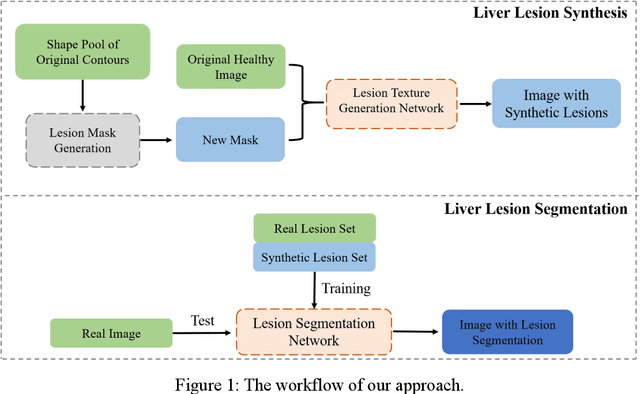
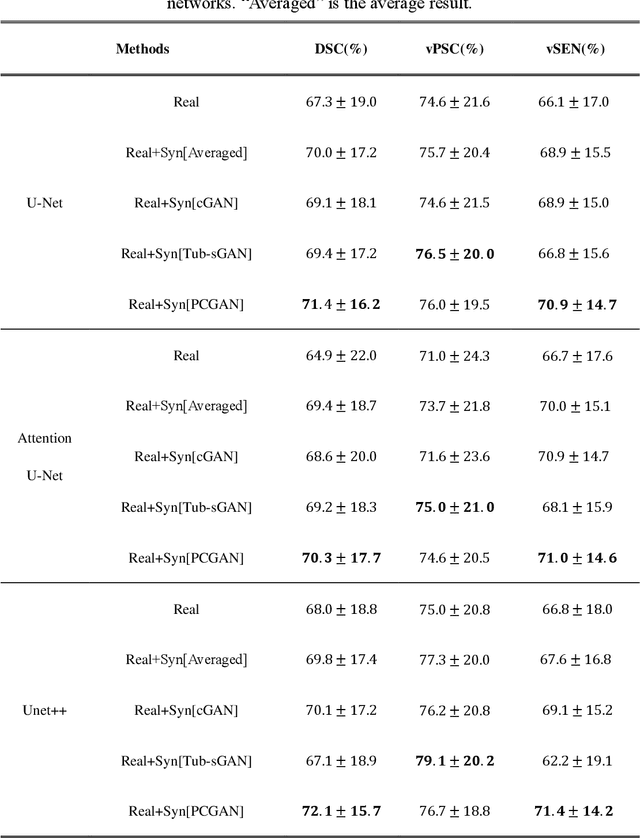
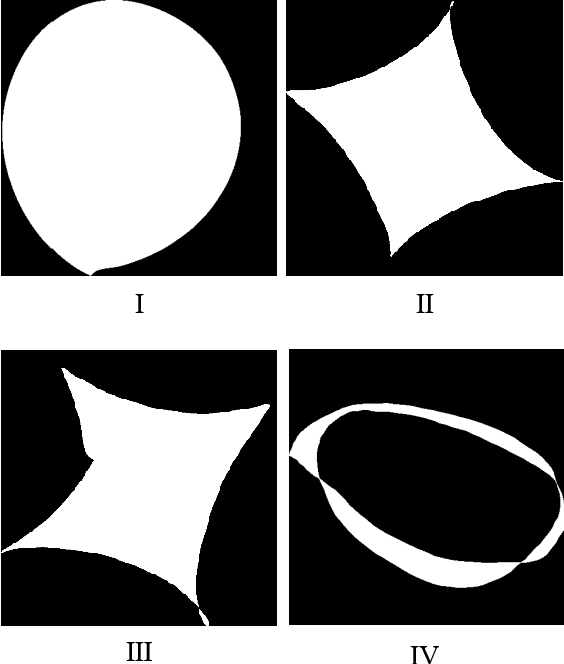
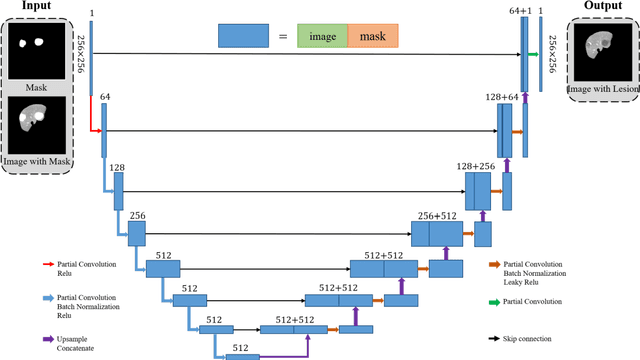
Automatic deep learning segmentation models has been shown to improve both the segmentation efficiency and the accuracy. However, training a robust segmentation model requires considerably large labeled training samples, which may be impractical. This study aimed to develop a deep learning framework for generating synthetic lesions that can be used to enhance network training. The lesion synthesis network is a modified generative adversarial network (GAN). Specifically, we innovated a partial convolution strategy to construct an Unet-like generator. The discriminator is designed using Wasserstein GAN with gradient penalty and spectral normalization. A mask generation method based on principal component analysis was developed to model various lesion shapes. The generated masks are then converted into liver lesions through a lesion synthesis network. The lesion synthesis framework was evaluated for lesion textures, and the synthetic lesions were used to train a lesion segmentation network to further validate the effectiveness of this framework. All the networks are trained and tested on the public dataset from LITS. The synthetic lesions generated by the proposed approach have very similar histogram distributions compared to the real lesions for the two employed texture parameters, GLCM-energy and GLCM-correlation. The Kullback-Leibler divergence of GLCM-energy and GLCM-correlation were 0.01 and 0.10, respectively. Including the synthetic lesions in the tumor segmentation network improved the segmentation dice performance of U-Net significantly from 67.3% to 71.4% (p<0.05). Meanwhile, the volume precision and sensitivity improve from 74.6% to 76.0% (p=0.23) and 66.1% to 70.9% (p<0.01), respectively. The synthetic data significantly improves the segmentation performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge