How Close Are We? Limitations and Progress of AI Models in Banff Lesion Scoring
Paper and Code
Oct 31, 2025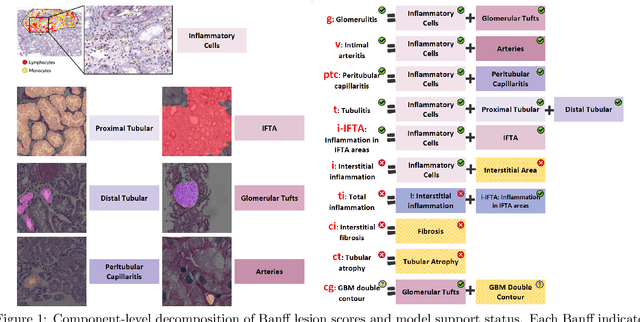
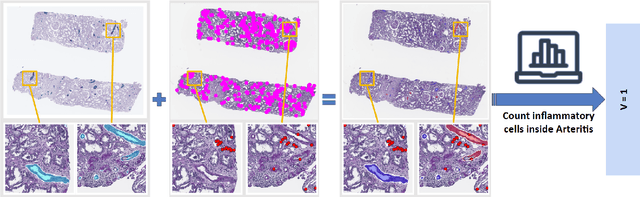
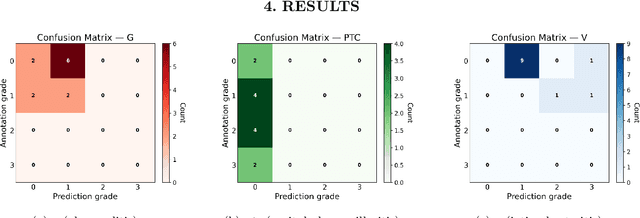
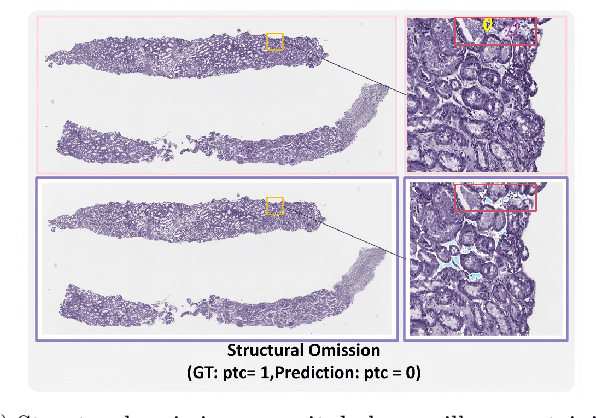
The Banff Classification provides the global standard for evaluating renal transplant biopsies, yet its semi-quantitative nature, complex criteria, and inter-observer variability present significant challenges for computational replication. In this study, we explore the feasibility of approximating Banff lesion scores using existing deep learning models through a modular, rule-based framework. We decompose each Banff indicator - such as glomerulitis (g), peritubular capillaritis (ptc), and intimal arteritis (v) - into its constituent structural and inflammatory components, and assess whether current segmentation and detection tools can support their computation. Model outputs are mapped to Banff scores using heuristic rules aligned with expert guidelines, and evaluated against expert-annotated ground truths. Our findings highlight both partial successes and critical failure modes, including structural omission, hallucination, and detection ambiguity. Even when final scores match expert annotations, inconsistencies in intermediate representations often undermine interpretability. These results reveal the limitations of current AI pipelines in replicating computational expert-level grading, and emphasize the importance of modular evaluation and computational Banff grading standard in guiding future model development for transplant pathology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge