Yinong Liu
Nonperiodic dynamic CT reconstruction using backward-warping INR with regularization of diffeomorphism (BIRD)
May 06, 2025



Abstract:Dynamic computed tomography (CT) reconstruction faces significant challenges in addressing motion artifacts, particularly for nonperiodic rapid movements such as cardiac imaging with fast heart rates. Traditional methods struggle with the extreme limited-angle problems inherent in nonperiodic cases. Deep learning methods have improved performance but face generalization challenges. Recent implicit neural representation (INR) techniques show promise through self-supervised deep learning, but have critical limitations: computational inefficiency due to forward-warping modeling, difficulty balancing DVF complexity with anatomical plausibility, and challenges in preserving fine details without additional patient-specific pre-scans. This paper presents a novel INR-based framework, BIRD, for nonperiodic dynamic CT reconstruction. It addresses these challenges through four key contributions: (1) backward-warping deformation that enables direct computation of each dynamic voxel with significantly reduced computational cost, (2) diffeomorphism-based DVF regularization that ensures anatomically plausible deformations while maintaining representational capacity, (3) motion-compensated analytical reconstruction that enhances fine details without requiring additional pre-scans, and (4) dimensional-reduction design for efficient 4D coordinate encoding. Through various simulations and practical studies, including digital and physical phantoms and retrospective patient data, we demonstrate the effectiveness of our approach for nonperiodic dynamic CT reconstruction with enhanced details and reduced motion artifacts. The proposed framework enables more accurate dynamic CT reconstruction with potential clinical applications, such as one-beat cardiac reconstruction, cinematic image sequences for functional imaging, and motion artifact reduction in conventional CT scans.
Investigation of domain gap problem in several deep-learning-based CT metal artefact reduction methods
Nov 25, 2021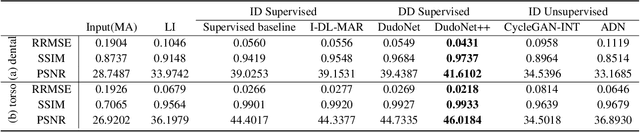
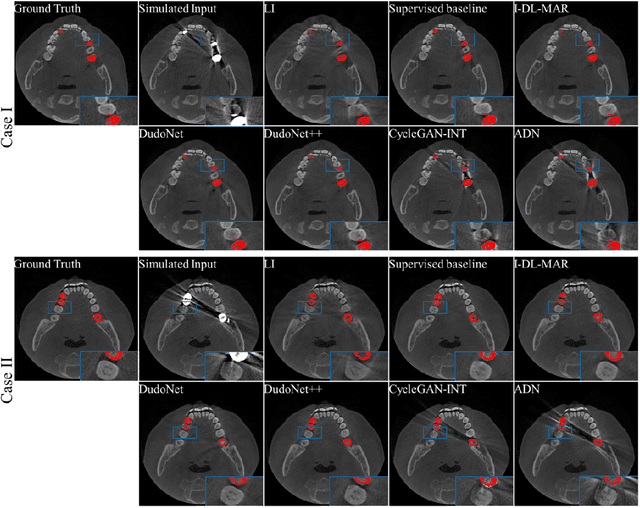
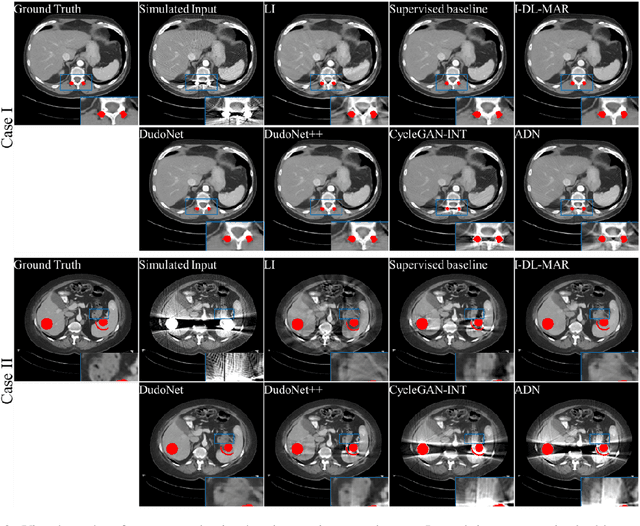
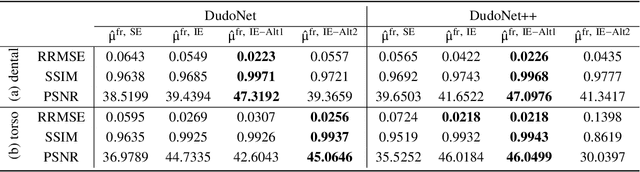
Abstract:Metal artefacts in CT images may disrupt image quality and interfere with diagnosis. Recently many deep-learning-based CT metal artefact reduction (MAR) methods have been proposed. Current deep MAR methods may be troubled with domain gap problem, where methods trained on simulated data cannot perform well on practical data. In this work, we experimentally investigate two image-domain supervised methods, two dual-domain supervised methods and two image-domain unsupervised methods on a dental dataset and a torso dataset, to explore whether domain gap problem exists or is overcome. We find that I-DL-MAR and DudoNet are effective for practical data of the torso dataset, indicating the domain gap problem is solved. However, none of the investigated methods perform satisfactorily on practical data of the dental dataset. Based on the experimental results, we further analyze the causes of domain gap problem for each method and dataset, which may be beneficial for improving existing methods or designing new ones. The findings suggest that the domain gap problem in deep MAR methods remains to be addressed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge