Yinling Qian
SAGS: Self-Adaptive Alias-Free Gaussian Splatting for Dynamic Surgical Endoscopic Reconstruction
Oct 31, 2025Abstract:Surgical reconstruction of dynamic tissues from endoscopic videos is a crucial technology in robot-assisted surgery. The development of Neural Radiance Fields (NeRFs) has greatly advanced deformable tissue reconstruction, achieving high-quality results from video and image sequences. However, reconstructing deformable endoscopic scenes remains challenging due to aliasing and artifacts caused by tissue movement, which can significantly degrade visualization quality. The introduction of 3D Gaussian Splatting (3DGS) has improved reconstruction efficiency by enabling a faster rendering pipeline. Nevertheless, existing 3DGS methods often prioritize rendering speed while neglecting these critical issues. To address these challenges, we propose SAGS, a self-adaptive alias-free Gaussian splatting framework. We introduce an attention-driven, dynamically weighted 4D deformation decoder, leveraging 3D smoothing filters and 2D Mip filters to mitigate artifacts in deformable tissue reconstruction and better capture the fine details of tissue movement. Experimental results on two public benchmarks, EndoNeRF and SCARED, demonstrate that our method achieves superior performance in all metrics of PSNR, SSIM, and LPIPS compared to the state of the art while also delivering better visualization quality.
Hepatic vessel segmentation based on 3D swin-transformer with inductive biased multi-head self-attention
Nov 22, 2021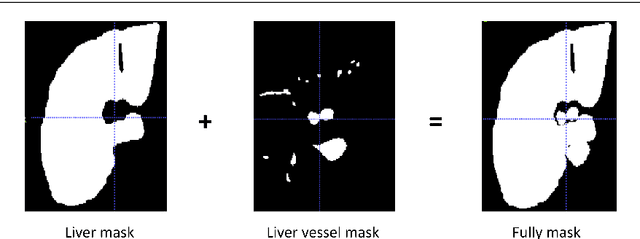
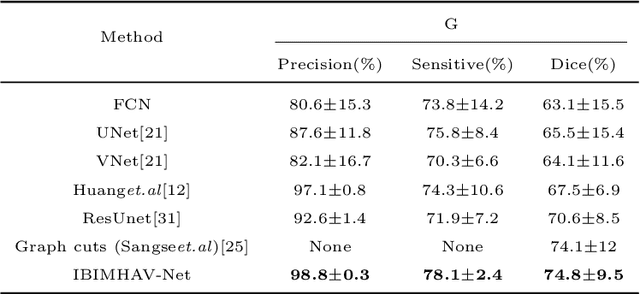
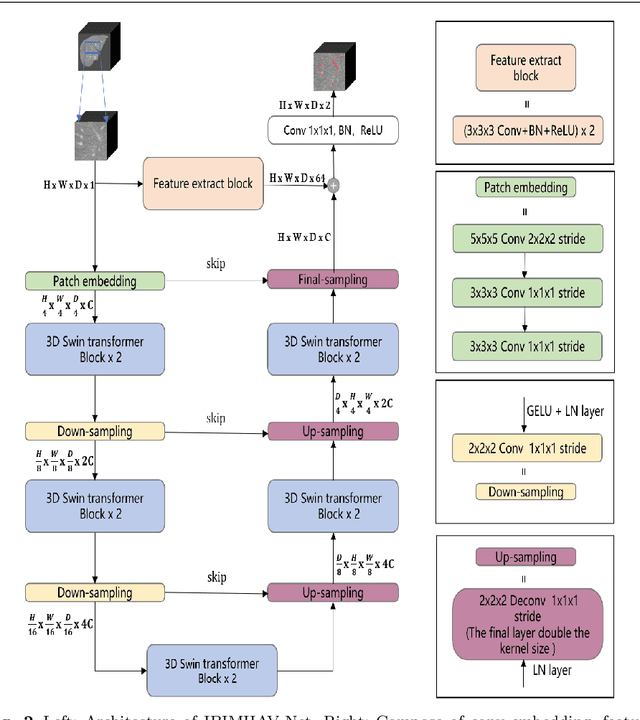
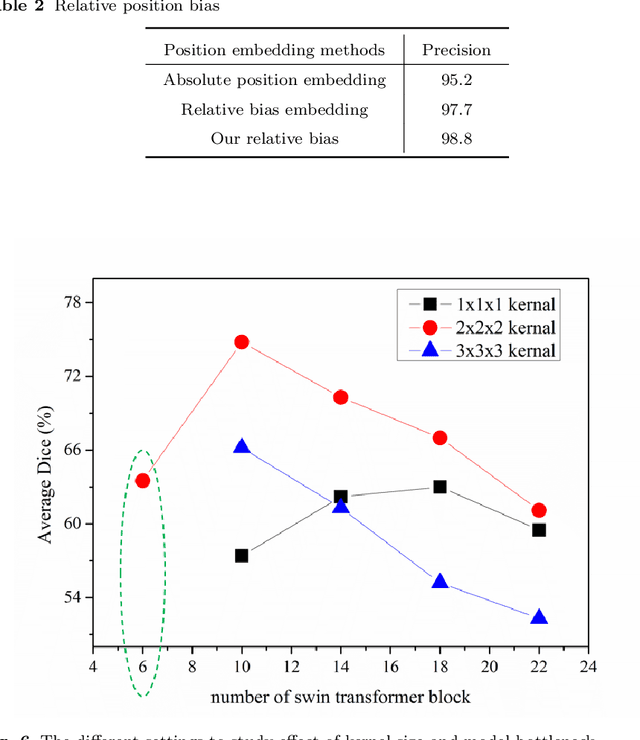
Abstract:Purpose: Segmentation of liver vessels from CT images is indispensable prior to surgical planning and aroused broad range of interests in the medical image analysis community. Due to the complex structure and low contrast background, automatic liver vessel segmentation remains particularly challenging. Most of the related researches adopt FCN, U-net, and V-net variants as a backbone. However, these methods mainly focus on capturing multi-scale local features which may produce misclassified voxels due to the convolutional operator's limited locality reception field. Methods: We propose a robust end-to-end vessel segmentation network called Inductive BIased Multi-Head Attention Vessel Net(IBIMHAV-Net) by expanding swin transformer to 3D and employing an effective combination of convolution and self-attention. In practice, we introduce the voxel-wise embedding rather than patch-wise embedding to locate precise liver vessel voxels, and adopt multi-scale convolutional operators to gain local spatial information. On the other hand, we propose the inductive biased multi-head self-attention which learns inductive biased relative positional embedding from initialized absolute position embedding. Based on this, we can gain a more reliable query and key matrix. To validate the generalization of our model, we test on samples which have different structural complexity. Results: We conducted experiments on the 3DIRCADb datasets. The average dice and sensitivity of the four tested cases were 74.8% and 77.5%, which exceed results of existing deep learning methods and improved graph cuts method. Conclusion: The proposed model IBIMHAV-Net provides an automatic, accurate 3D liver vessel segmentation with an interleaved architecture that better utilizes both global and local spatial features in CT volumes. It can be further extended for other clinical data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge