Yanjun Li
Nankai University
Tableaux for the Logic of Strategically Knowing How
Jul 11, 2023
Abstract:The logic of goal-directed knowing-how extends the standard epistemic logic with an operator of knowing-how. The knowing-how operator is interpreted as that there exists a strategy such that the agent knows that the strategy can make sure that p. This paper presents a tableau procedure for the multi-agent version of the logic of strategically knowing-how and shows the soundness and completeness of this tableau procedure. This paper also shows that the satisfiability problem of the logic can be decided in PSPACE.
* In Proceedings TARK 2023, arXiv:2307.04005
A Bioinspired Synthetic Nervous System Controller for Pick-and-Place Manipulation
May 18, 2023



Abstract:The Synthetic Nervous System (SNS) is a biologically inspired neural network (NN). Due to its capability of capturing complex mechanisms underlying neural computation, an SNS model is a candidate for building compact and interpretable NN controllers for robots. Previous work on SNSs has focused on applying the model to the control of legged robots and the design of functional subnetworks (FSNs) to realize dynamical systems. However, the FSN approach has previously relied on the analytical solution of the governing equations, which is difficult for designing more complex NN controllers. Incorporating plasticity into SNSs and using learning algorithms to tune the parameters offers a promising solution for systematic design in this situation. In this paper, we theoretically analyze the computational advantages of SNSs compared with other classical artificial neural networks. We then use learning algorithms to develop compact subnetworks for implementing addition, subtraction, division, and multiplication. We also combine the learning-based methodology with a bioinspired architecture to design an interpretable SNS for the pick-and-place control of a simulated gantry system. Finally, we show that the SNS controller is successfully transferred to a real-world robotic platform without further tuning of the parameters, verifying the effectiveness of our approach.
BatmanNet: Bi-branch Masked Graph Transformer Autoencoder for Molecular Representation
Nov 29, 2022



Abstract:Although substantial efforts have been made using graph neural networks (GNNs) for AI-driven drug discovery (AIDD), effective molecular representation learning remains an open challenge, especially in the case of insufficient labeled molecules. Recent studies suggest that big GNN models pre-trained by self-supervised learning on unlabeled datasets enable better transfer performance in downstream molecular property prediction tasks. However, they often require large-scale datasets and considerable computational resources, which is time-consuming, computationally expensive, and environmentally unfriendly. To alleviate these limitations, we propose a novel pre-training model for molecular representation learning, Bi-branch Masked Graph Transformer Autoencoder (BatmanNet). BatmanNet features two tailored and complementary graph autoencoders to reconstruct the missing nodes and edges from a masked molecular graph. To our surprise, BatmanNet discovered that the highly masked proportion (60%) of the atoms and bonds achieved the best performance. We further propose an asymmetric graph-based encoder-decoder architecture for either nodes and edges, where a transformer-based encoder only takes the visible subset of nodes or edges, and a lightweight decoder reconstructs the original molecule from the latent representation and mask tokens. With this simple yet effective asymmetrical design, our BatmanNet can learn efficiently even from a much smaller-scale unlabeled molecular dataset to capture the underlying structural and semantic information, overcoming a major limitation of current deep neural networks for molecular representation learning. For instance, using only 250K unlabelled molecules as pre-training data, our BatmanNet with 2.575M parameters achieves a 0.5% improvement on the average AUC compared with the current state-of-the-art method with 100M parameters pre-trained on 11M molecules.
Knowing How to Plan
Jun 22, 2021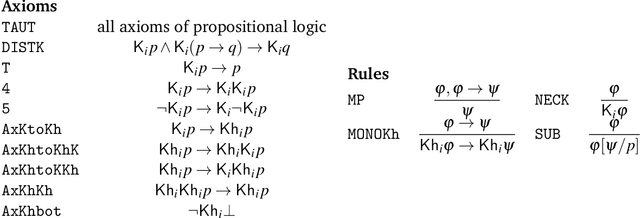
Abstract:Various planning-based know-how logics have been studied in the recent literature. In this paper, we use such a logic to do know-how-based planning via model checking. In particular, we can handle the higher-order epistemic planning involving know-how formulas as the goal, e.g., find a plan to make sure p such that the adversary does not know how to make p false in the future. We give a PTIME algorithm for the model checking problem over finite epistemic transition systems and axiomatize the logic under the assumption of perfect recall.
* In Proceedings TARK 2021, arXiv:2106.10886
Joint Dimensionality Reduction for Separable Embedding Estimation
Jan 14, 2021
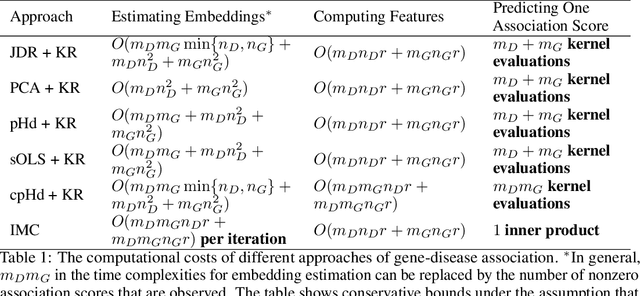
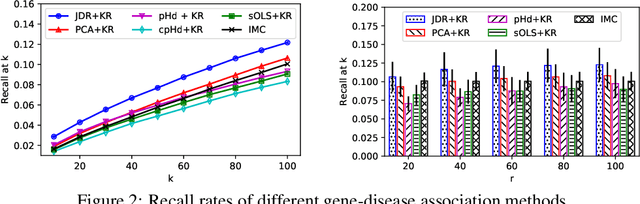
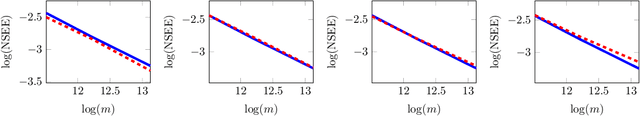
Abstract:Low-dimensional embeddings for data from disparate sources play critical roles in multi-modal machine learning, multimedia information retrieval, and bioinformatics. In this paper, we propose a supervised dimensionality reduction method that learns linear embeddings jointly for two feature vectors representing data of different modalities or data from distinct types of entities. We also propose an efficient feature selection method that complements, and can be applied prior to, our joint dimensionality reduction method. Assuming that there exist true linear embeddings for these features, our analysis of the error in the learned linear embeddings provides theoretical guarantees that the dimensionality reduction method accurately estimates the true embeddings when certain technical conditions are satisfied and the number of samples is sufficiently large. The derived sample complexity results are echoed by numerical experiments. We apply the proposed dimensionality reduction method to gene-disease association, and predict unknown associations using kernel regression on the dimension-reduced feature vectors. Our approach compares favorably against other dimensionality reduction methods, and against a state-of-the-art method of bilinear regression for predicting gene-disease associations.
PRI-VAE: Principle-of-Relevant-Information Variational Autoencoders
Jul 13, 2020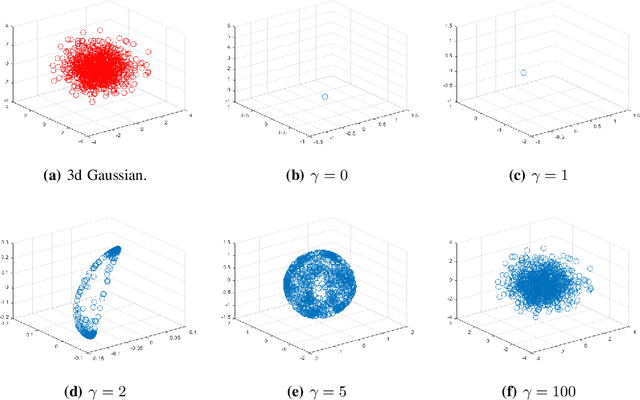
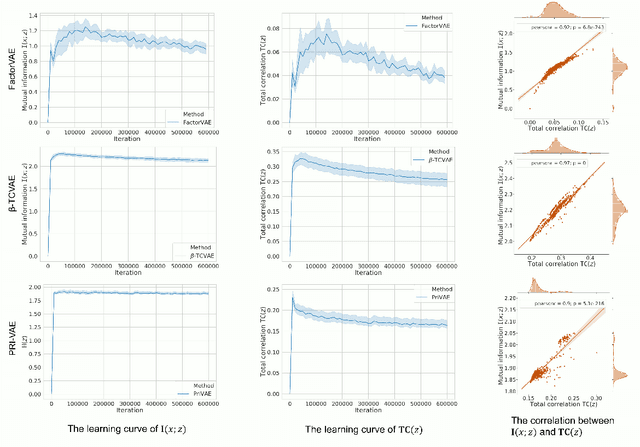
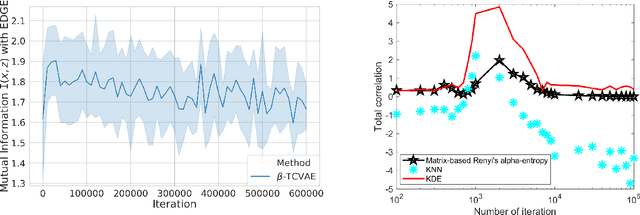
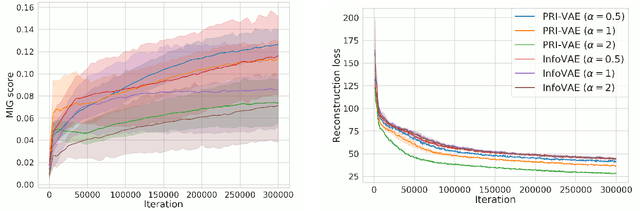
Abstract:Although substantial efforts have been made to learn disentangled representations under the variational autoencoder (VAE) framework, the fundamental properties to the dynamics of learning of most VAE models still remain unknown and under-investigated. In this work, we first propose a novel learning objective, termed the principle-of-relevant-information variational autoencoder (PRI-VAE), to learn disentangled representations. We then present an information-theoretic perspective to analyze existing VAE models by inspecting the evolution of some critical information-theoretic quantities across training epochs. Our observations unveil some fundamental properties associated with VAEs. Empirical results also demonstrate the effectiveness of PRI-VAE on four benchmark data sets.
Application of Deep Interpolation Network for Clustering of Physiologic Time Series
Apr 27, 2020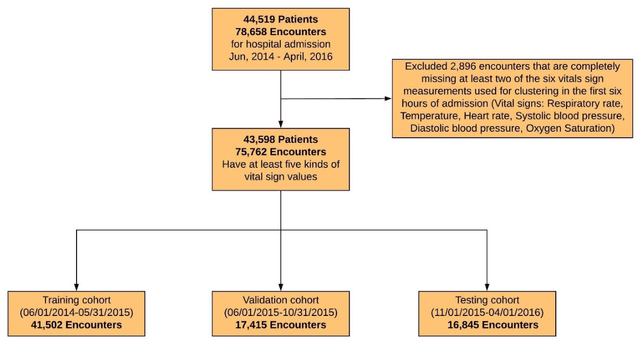
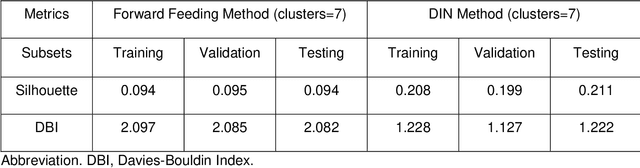
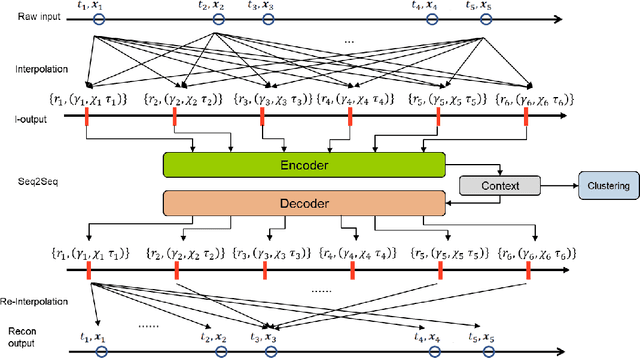
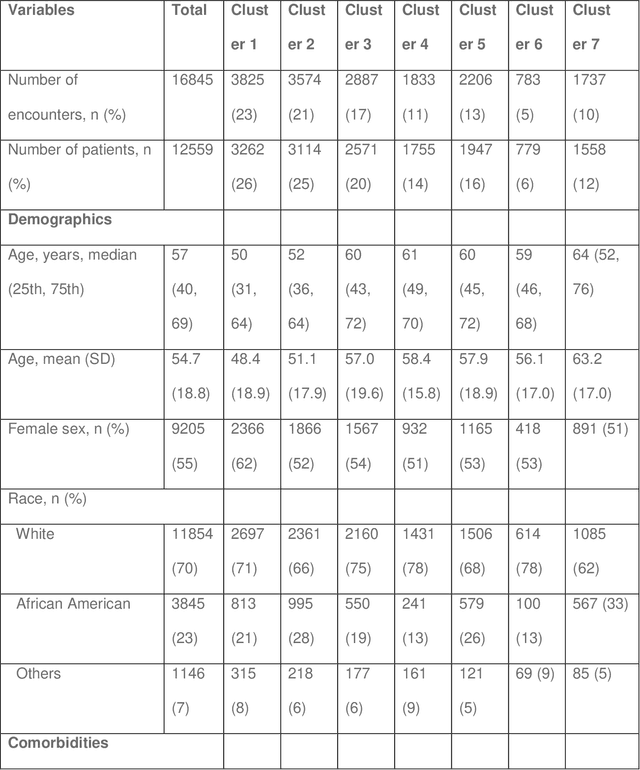
Abstract:Background: During the early stages of hospital admission, clinicians must use limited information to make diagnostic and treatment decisions as patient acuity evolves. However, it is common that the time series vital sign information from patients to be both sparse and irregularly collected, which poses a significant challenge for machine / deep learning techniques to analyze and facilitate the clinicians to improve the human health outcome. To deal with this problem, We propose a novel deep interpolation network to extract latent representations from sparse and irregularly sampled time-series vital signs measured within six hours of hospital admission. Methods: We created a single-center longitudinal dataset of electronic health record data for all (n=75,762) adult patient admissions to a tertiary care center lasting six hours or longer, using 55% of the dataset for training, 23% for validation, and 22% for testing. All raw time series within six hours of hospital admission were extracted for six vital signs (systolic blood pressure, diastolic blood pressure, heart rate, temperature, blood oxygen saturation, and respiratory rate). A deep interpolation network is proposed to learn from such irregular and sparse multivariate time series data to extract the fixed low-dimensional latent patterns. We use k-means clustering algorithm to clusters the patient admissions resulting into 7 clusters. Findings: Training, validation, and testing cohorts had similar age (55-57 years), sex (55% female), and admission vital signs. Seven distinct clusters were identified. M Interpretation: In a heterogeneous cohort of hospitalized patients, a deep interpolation network extracted representations from vital sign data measured within six hours of hospital admission. This approach may have important implications for clinical decision-support under time constraints and uncertainty.
A Set-Theoretic Study of the Relationships of Image Models and Priors for Restoration Problems
Mar 29, 2020



Abstract:Image prior modeling is the key issue in image recovery, computational imaging, compresses sensing, and other inverse problems. Recent algorithms combining multiple effective priors such as the sparse or low-rank models, have demonstrated superior performance in various applications. However, the relationships among the popular image models are unclear, and no theory in general is available to demonstrate their connections. In this paper, we present a theoretical analysis on the image models, to bridge the gap between applications and image prior understanding, including sparsity, group-wise sparsity, joint sparsity, and low-rankness, etc. We systematically study how effective each image model is for image restoration. Furthermore, we relate the denoising performance improvement by combining multiple models, to the image model relationships. Extensive experiments are conducted to compare the denoising results which are consistent with our analysis. On top of the model-based methods, we quantitatively demonstrate the image properties that are inexplicitly exploited by deep learning method, of which can further boost the denoising performance by combining with its complementary image models.
DeepAtom: A Framework for Protein-Ligand Binding Affinity Prediction
Dec 01, 2019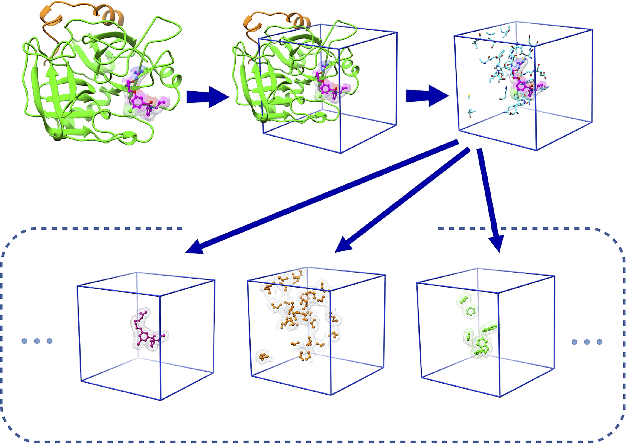
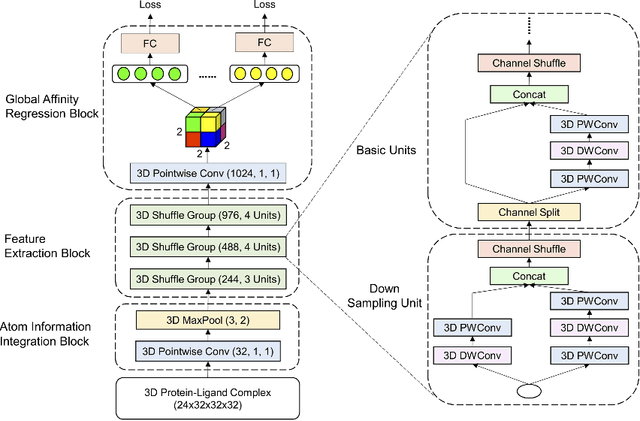
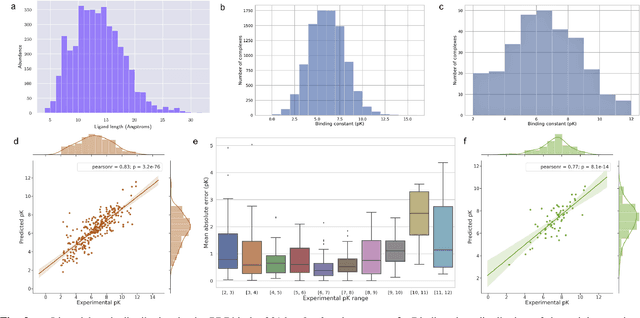
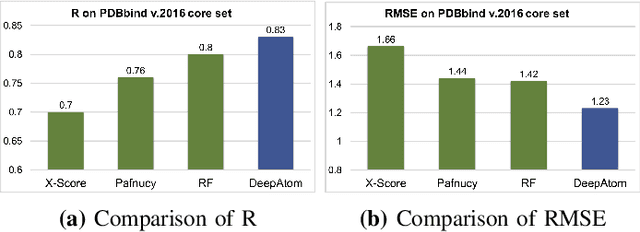
Abstract:The cornerstone of computational drug design is the calculation of binding affinity between two biological counterparts, especially a chemical compound, i.e., a ligand, and a protein. Predicting the strength of protein-ligand binding with reasonable accuracy is critical for drug discovery. In this paper, we propose a data-driven framework named DeepAtom to accurately predict the protein-ligand binding affinity. With 3D Convolutional Neural Network (3D-CNN) architecture, DeepAtom could automatically extract binding related atomic interaction patterns from the voxelized complex structure. Compared with the other CNN based approaches, our light-weight model design effectively improves the model representational capacity, even with the limited available training data. With validation experiments on the PDBbind v.2016 benchmark and the independent Astex Diverse Set, we demonstrate that the less feature engineering dependent DeepAtom approach consistently outperforms the other state-of-the-art scoring methods. We also compile and propose a new benchmark dataset to further improve the model performances. With the new dataset as training input, DeepAtom achieves Pearson's R=0.83 and RMSE=1.23 pK units on the PDBbind v.2016 core set. The promising results demonstrate that DeepAtom models can be potentially adopted in computational drug development protocols such as molecular docking and virtual screening.
* Accepted IEEE BIBM 2019 paper with minor revisions
Algorithmic Design and Implementation of Unobtrusive Multistatic Serial LiDAR Image
Nov 08, 2019
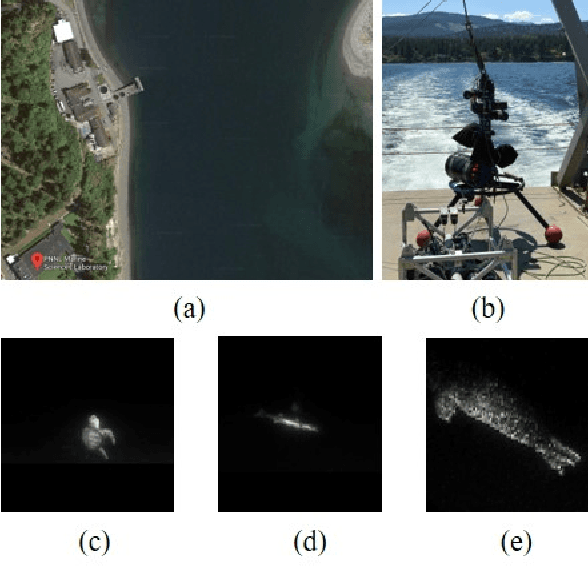
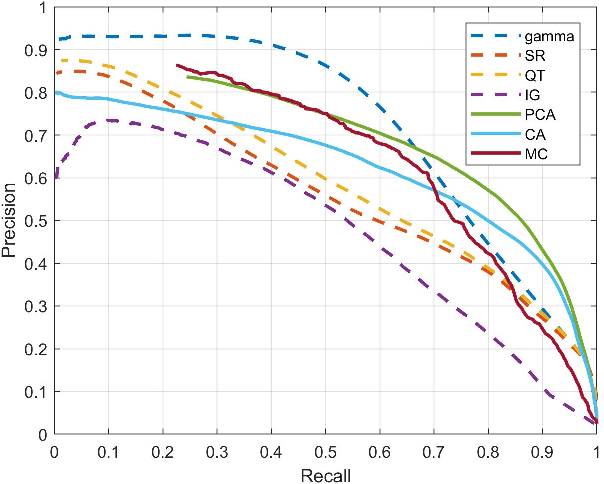
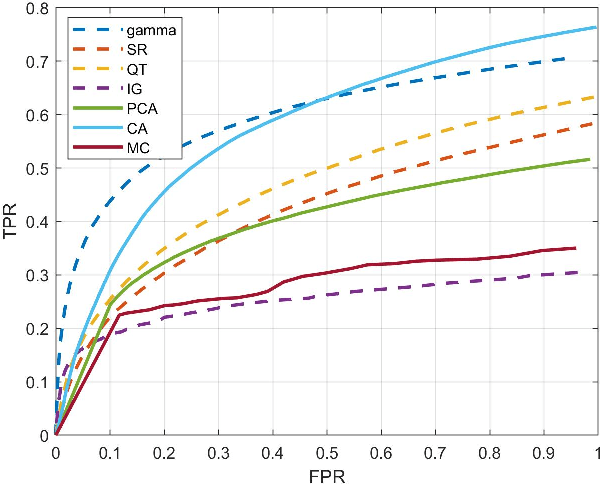
Abstract:To fully understand interactions between marine hydrokinetic (MHK) equipment and marine animals, a fast and effective monitoring system is required to capture relevant information whenever underwater animals appear. A new automated underwater imaging system composed of LiDAR (Light Detection and Ranging) imaging hardware and a scene understanding software module named Unobtrusive Multistatic Serial LiDAR Imager (UMSLI) to supervise the presence of animals near turbines. UMSLI integrates the front end LiDAR hardware and a series of software modules to achieve image preprocessing, detection, tracking, segmentation and classification in a hierarchical manner.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge