Xiaotong Jiang
Oral Imaging for Malocclusion Issues Assessments: OMNI Dataset, Deep Learning Baselines and Benchmarking
May 21, 2025Abstract:Malocclusion is a major challenge in orthodontics, and its complex presentation and diverse clinical manifestations make accurate localization and diagnosis particularly important. Currently, one of the major shortcomings facing the field of dental image analysis is the lack of large-scale, accurately labeled datasets dedicated to malocclusion issues, which limits the development of automated diagnostics in the field of dentistry and leads to a lack of diagnostic accuracy and efficiency in clinical practice. Therefore, in this study, we propose the Oral and Maxillofacial Natural Images (OMNI) dataset, a novel and comprehensive dental image dataset aimed at advancing the study of analyzing dental images for issues of malocclusion. Specifically, the dataset contains 4166 multi-view images with 384 participants in data collection and annotated by professional dentists. In addition, we performed a comprehensive validation of the created OMNI dataset, including three CNN-based methods, two Transformer-based methods, and one GNN-based method, and conducted automated diagnostic experiments for malocclusion issues. The experimental results show that the OMNI dataset can facilitate the automated diagnosis research of malocclusion issues and provide a new benchmark for the research in this field. Our OMNI dataset and baseline code are publicly available at https://github.com/RoundFaceJ/OMNI.
Opinion Tree Parsing for Aspect-based Sentiment Analysis
Jun 15, 2023Abstract:Extracting sentiment elements using pre-trained generative models has recently led to large improvements in aspect-based sentiment analysis benchmarks. However, these models always need large-scale computing resources, and they also ignore explicit modeling of structure between sentiment elements. To address these challenges, we propose an opinion tree parsing model, aiming to parse all the sentiment elements from an opinion tree, which is much faster, and can explicitly reveal a more comprehensive and complete aspect-level sentiment structure. In particular, we first introduce a novel context-free opinion grammar to normalize the opinion tree structure. We then employ a neural chart-based opinion tree parser to fully explore the correlations among sentiment elements and parse them into an opinion tree structure. Extensive experiments show the superiority of our proposed model and the capacity of the opinion tree parser with the proposed context-free opinion grammar. More importantly, the results also prove that our model is much faster than previous models.
Technical Background for "A Precision Medicine Approach to Develop and Internally Validate Optimal Exercise and Weight Loss Treatments for Overweight and Obese Adults with Knee Osteoarthritis"
Feb 20, 2020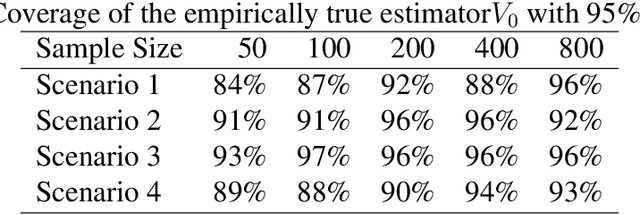
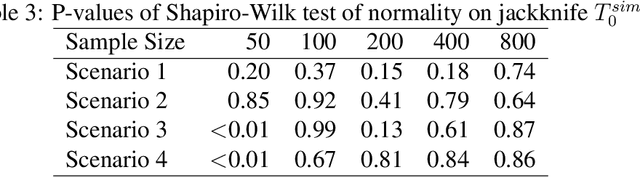
Abstract:We provide additional statistical background for the methodology developed in the clinical analysis of knee osteoarthritis in "A Precision Medicine Approach to Develop and Internally Validate Optimal Exercise and Weight Loss Treatments for Overweight and Obese Adults with Knee Osteoarthritis" (Jiang et al. 2020). Jiang et al. 2020 proposed a pipeline to learn optimal treatment rules with precision medicine models and compared them with zero-order models with a Z-test. The model performance was based on value functions, a scalar that predicts the future reward of each decision rule. The jackknife (i.e., leave-one-out cross validation) method was applied to estimate the value function and its variance of several outcomes in IDEA. IDEA is a randomized clinical trial studying three interventions (exercise (E), dietary weight loss (D), and D+E) on overweight and obese participants with knee osteoarthritis. In this report, we expand the discussion and justification with additional statistical background. We elaborate more on the background of precision medicine, the derivation of the jackknife estimator of value function and its estimated variance, the consistency property of jackknife estimator, as well as additional simulation results that reflect more of the performance of jackknife estimators. We recommend reading Jiang et al. 2020 for clinical application and interpretation of the optimal ITR of knee osteoarthritis as well as the overall understanding of the pipeline and recommend using this article to understand the underlying statistical derivation and methodology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge