Xiaohua Qian
Generalizable Pancreas Segmentation via a Dual Self-Supervised Learning Framework
May 12, 2025Abstract:Recently, numerous pancreas segmentation methods have achieved promising performance on local single-source datasets. However, these methods don't adequately account for generalizability issues, and hence typically show limited performance and low stability on test data from other sources. Considering the limited availability of distinct data sources, we seek to improve the generalization performance of a pancreas segmentation model trained with a single-source dataset, i.e., the single source generalization task. In particular, we propose a dual self-supervised learning model that incorporates both global and local anatomical contexts. Our model aims to fully exploit the anatomical features of the intra-pancreatic and extra-pancreatic regions, and hence enhance the characterization of the high-uncertainty regions for more robust generalization. Specifically, we first construct a global-feature contrastive self-supervised learning module that is guided by the pancreatic spatial structure. This module obtains complete and consistent pancreatic features through promoting intra-class cohesion, and also extracts more discriminative features for differentiating between pancreatic and non-pancreatic tissues through maximizing inter-class separation. It mitigates the influence of surrounding tissue on the segmentation outcomes in high-uncertainty regions. Subsequently, a local-image restoration self-supervised learning module is introduced to further enhance the characterization of the high uncertainty regions. In this module, informative anatomical contexts are actually learned to recover randomly corrupted appearance patterns in those regions.
A Dual-Task Synergy-Driven Generalization Framework for Pancreatic Cancer Segmentation in CT Scans
May 03, 2025Abstract:Pancreatic cancer, characterized by its notable prevalence and mortality rates, demands accurate lesion delineation for effective diagnosis and therapeutic interventions. The generalizability of extant methods is frequently compromised due to the pronounced variability in imaging and the heterogeneous characteristics of pancreatic lesions, which may mimic normal tissues and exhibit significant inter-patient variability. Thus, we propose a generalization framework that synergizes pixel-level classification and regression tasks, to accurately delineate lesions and improve model stability. This framework not only seeks to align segmentation contours with actual lesions but also uses regression to elucidate spatial relationships between diseased and normal tissues, thereby improving tumor localization and morphological characterization. Enhanced by the reciprocal transformation of task outputs, our approach integrates additional regression supervision within the segmentation context, bolstering the model's generalization ability from a dual-task perspective. Besides, dual self-supervised learning in feature spaces and output spaces augments the model's representational capability and stability across different imaging views. Experiments on 594 samples composed of three datasets with significant imaging differences demonstrate that our generalized pancreas segmentation results comparable to mainstream in-domain validation performance (Dice: 84.07%). More importantly, it successfully improves the results of the highly challenging cross-lesion generalized pancreatic cancer segmentation task by 9.51%. Thus, our model constitutes a resilient and efficient foundational technological support for pancreatic disease management and wider medical applications. The codes will be released at https://github.com/SJTUBME-QianLab/Dual-Task-Seg.
Probability Map Guided Bi-directional Recurrent UNet for Pancreas Segmentation
Apr 07, 2019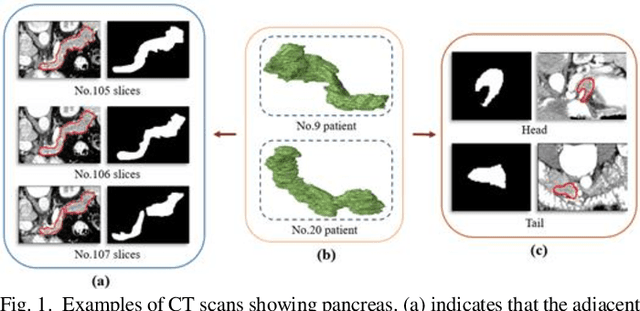
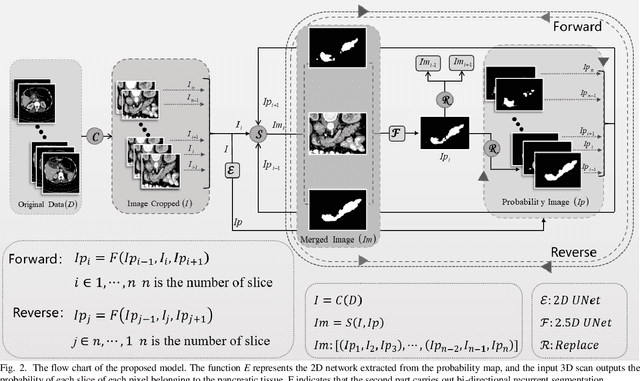
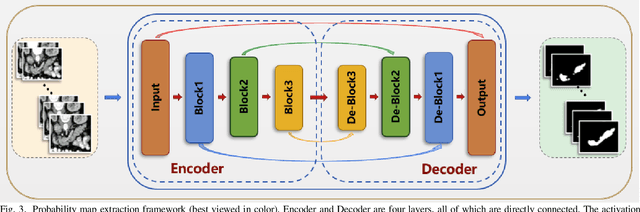
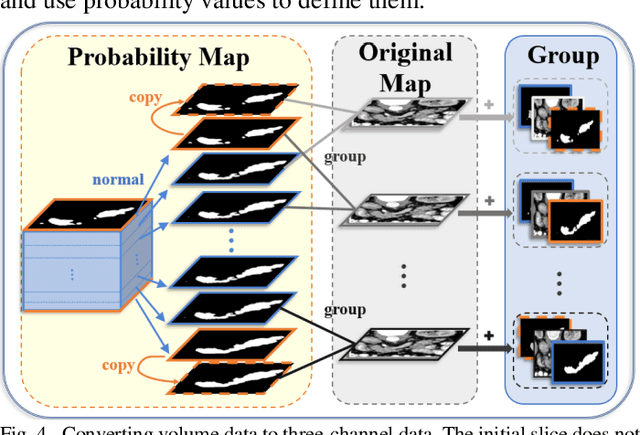
Abstract:Pancreatic cancer is one of the most lethal cancers as morbidity approximates mortality. A method for accurately segmenting the pancreas can assist doctors in the diagnosis and treatment of pancreatic cancer, but huge differences in shape and volume bring difficulties in segmentation. Among the current widely used approaches, the 2D method ignores the spatial information, and the 3D model is limited by high resource consumption and GPU memory occupancy. To address these issues, we propose a bi-directional recurrent UNet based on probabilistic map guidance (PBR-UNet). PBR-UNet includes a feature extraction module for extracting pixel-level probabilistic maps and a bi-directional recurrent module for fine segmentation. The extracted probabilistic map will be used to guide the fine segmentation and bi-directional recurrent module integrates contextual information into the entire network to avoid the loss of spatial information in propagation. By combining the probabilistic map of the adjacent slices with the bi-directional recurrent segmentation of intermediary slice, this paper solves the problem that the 2D network loses three-dimensional information and the 3D model leads to large computational resource consumption. We used Dice similarity coefficients (DSC) to evaluate our approach on NIH pancreatic datasets and eventually achieved a competitive result of 83.35%.
Pancreas Segmentation via Spatial Context based U-net and Bidirectional LSTM
Mar 03, 2019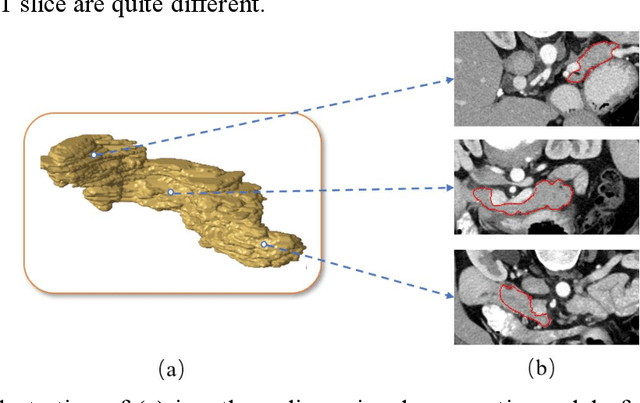
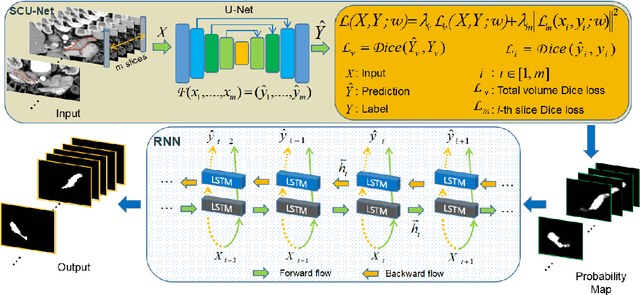
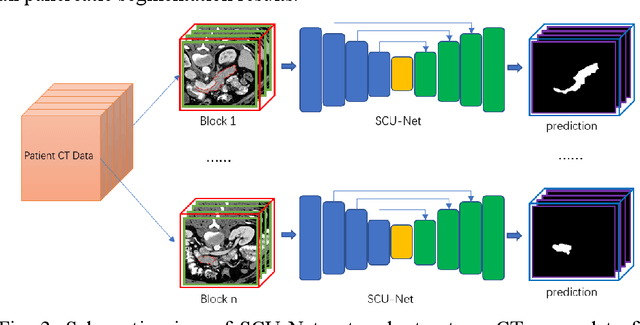
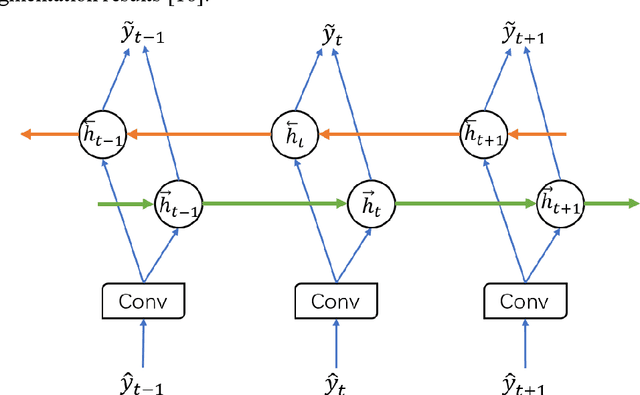
Abstract:Pancreas is characterized by small size and irregular shape, so achieving accurate pancreas segmentation is challenging. Traditional 2D pancreas segmentation network based on the independent 2D image slices, which often leads to spatial discontinuity problem. Therefore, how to utility spatial context information is the key point to improve the segmentation quality. In this paper, we proposed a divide-and-conquer strategy, divided the abdominal CT scans into several isometric blocks. And we designed a multiple channels convolutional neural network to learn the local spatial context characteristics from blocks called SCU-Net. SCU-Net is a partial 3D segmentation idea, which transforms overall pancreas segmentation into a combination of multiple local segmentation results. In order to improve the segmentation accuracy for each layer, we also proposed a new loss function for inter-slice constrain and regularization. Thereafter, we introduced the BiCLSTM network for stimulating the interaction between bidirectional segmentation sequence, thus making up the boundary defect and fault problem of the segmentation results. We trained SCU-Net+BiLSTM network respectively, and evaluated segmentation result on the NIH data set. Keywords: Pancreas Segmentation, Convolutional Neural Networks, Recurrent Neural Networks, Deep Learning, Inter-slice Regularization
DC-Al GAN: Pseudoprogression and True Tumor Progression of Glioblastoma multiform Image Classification Based On DCGAN and Alexnet
Feb 26, 2019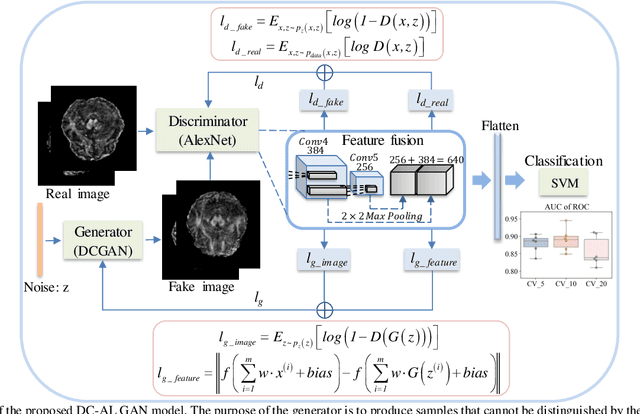
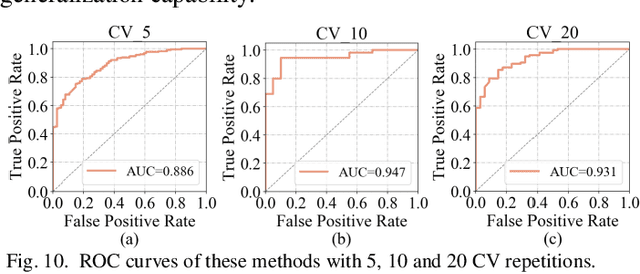
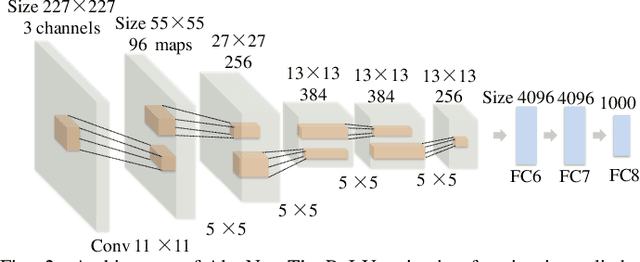
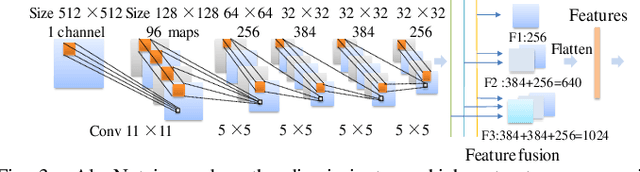
Abstract:Glioblastoma multiform (GBM) is a kind of head tumor with an extraordinarily complex treatment process. The survival period is typically 14-16 months, and the 2 year survival rate is approximately 26%-33%. The clinical treatment strategies for the pseudoprogression (PsP) and true tumor progression (TTP) of GBM are different, so accurately distinguishing these two conditions is particularly significant.As PsP and TTP of GBM are similar in shape and other characteristics, it is hard to distinguish these two forms with precision. In order to differentiate them accurately, this paper introduces a feature learning method based on a generative adversarial network: DC-Al GAN. GAN consists of two architectures: generator and discriminator. Alexnet is used as the discriminator in this work. Owing to the adversarial and competitive relationship between generator and discriminator, the latter extracts highly concise features during training. In DC-Al GAN, features are extracted from Alexnet in the final classification phase, and the highly nature of them contributes positively to the classification accuracy.The generator in DC-Al GAN is modified by the deep convolutional generative adversarial network (DCGAN) by adding three convolutional layers. This effectively generates higher resolution sample images. Feature fusion is used to combine high layer features with low layer features, allowing for the creation and use of more precise features for classification. The experimental results confirm that DC-Al GAN achieves high accuracy on GBM datasets for PsP and TTP image classification, which is superior to other state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge