Weimiao Yu
Exploiting Layer Normalization Fine-tuning in Visual Transformer Foundation Models for Classification
Aug 11, 2025Abstract:LayerNorm is pivotal in Vision Transformers (ViTs), yet its fine-tuning dynamics under data scarcity and domain shifts remain underexplored. This paper shows that shifts in LayerNorm parameters after fine-tuning (LayerNorm shifts) are indicative of the transitions between source and target domains; its efficacy is contingent upon the degree to which the target training samples accurately represent the target domain, as quantified by our proposed Fine-tuning Shift Ratio ($FSR$). Building on this, we propose a simple yet effective rescaling mechanism using a scalar $\lambda$ that is negatively correlated to $FSR$ to align learned LayerNorm shifts with those ideal shifts achieved under fully representative data, combined with a cyclic framework that further enhances the LayerNorm fine-tuning. Extensive experiments across natural and pathological images, in both in-distribution (ID) and out-of-distribution (OOD) settings, and various target training sample regimes validate our framework. Notably, OOD tasks tend to yield lower $FSR$ and higher $\lambda$ in comparison to ID cases, especially with scarce data, indicating under-represented target training samples. Moreover, ViTFs fine-tuned on pathological data behave more like ID settings, favoring conservative LayerNorm updates. Our findings illuminate the underexplored dynamics of LayerNorm in transfer learning and provide practical strategies for LayerNorm fine-tuning.
Regional Registration of Whole Slide Image Stacks Containing Highly Deformed Artefacts
Feb 28, 2020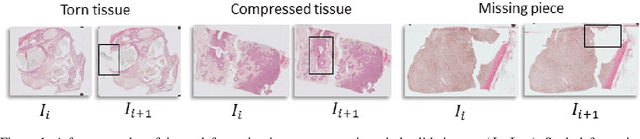
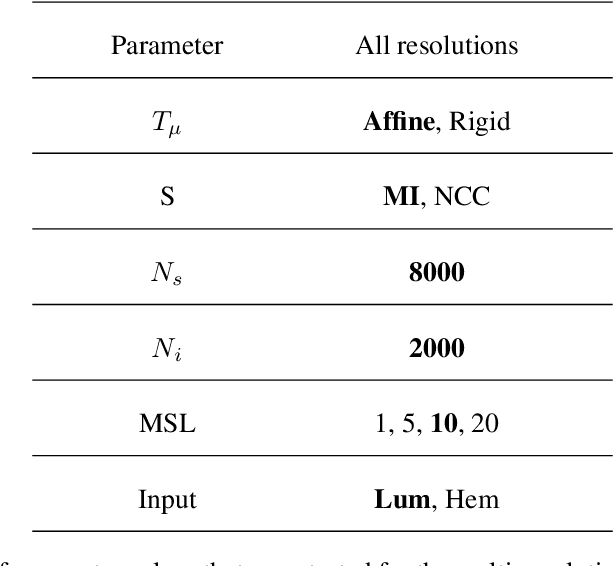
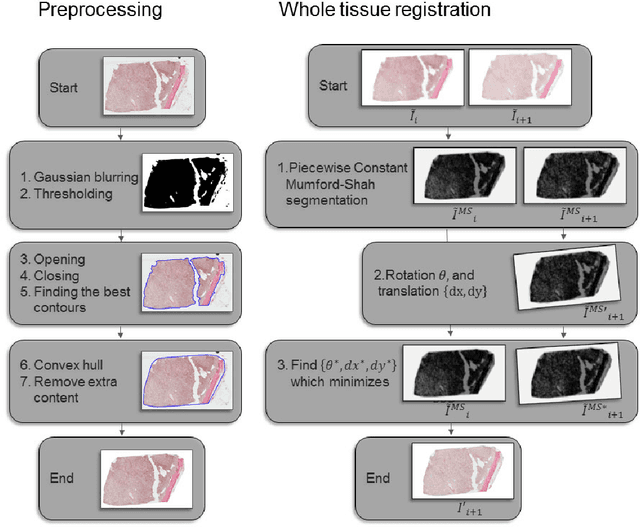
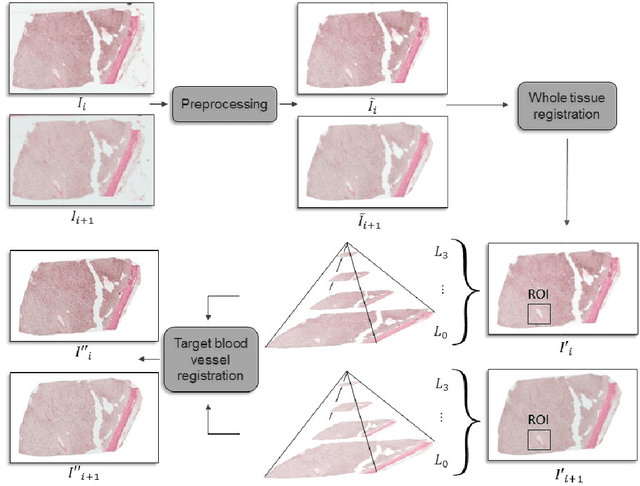
Abstract:Motivation: High resolution 2D whole slide imaging provides rich information about the tissue structure. This information can be a lot richer if these 2D images can be stacked into a 3D tissue volume. A 3D analysis, however, requires accurate reconstruction of the tissue volume from the 2D image stack. This task is not trivial due to the distortions that each individual tissue slice experiences while cutting and mounting the tissue on the glass slide. Performing registration for the whole tissue slices may be adversely affected by the deformed tissue regions. Consequently, regional registration is found to be more effective. In this paper, we propose an accurate and robust regional registration algorithm for whole slide images which incrementally focuses registration on the area around the region of interest. Results: Using mean similarity index as the metric, the proposed algorithm (mean $\pm$ std: $0.84 \pm 0.11$) followed by a fine registration algorithm ($0.86 \pm 0.08$) outperformed the state-of-the-art linear whole tissue registration algorithm ($0.74 \pm 0.19$) and the regional version of this algorithm ($0.81 \pm 0.15$). The proposed algorithm also outperforms the state-of-the-art nonlinear registration algorithm (original : $0.82 \pm 0.12$, regional : $0.77 \pm 0.22$) for whole slide images and a recently proposed patch-based registration algorithm (patch size 256: $0.79 \pm 0.16$ , patch size 512: $0.77 \pm 0.16$) for medical images. Availability: The C++ implementation code is available online at the github repository: https://github.com/MahsaPaknezhad/WSIRegistration
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge