Wah Chiu
MicroVQA: A Multimodal Reasoning Benchmark for Microscopy-Based Scientific Research
Mar 17, 2025Abstract:Scientific research demands sophisticated reasoning over multimodal data, a challenge especially prevalent in biology. Despite recent advances in multimodal large language models (MLLMs) for AI-assisted research, existing multimodal reasoning benchmarks only target up to college-level difficulty, while research-level benchmarks emphasize lower-level perception, falling short of the complex multimodal reasoning needed for scientific discovery. To bridge this gap, we introduce MicroVQA, a visual-question answering (VQA) benchmark designed to assess three reasoning capabilities vital in research workflows: expert image understanding, hypothesis generation, and experiment proposal. MicroVQA consists of 1,042 multiple-choice questions (MCQs) curated by biology experts across diverse microscopy modalities, ensuring VQA samples represent real scientific practice. In constructing the benchmark, we find that standard MCQ generation methods induce language shortcuts, motivating a new two-stage pipeline: an optimized LLM prompt structures question-answer pairs into MCQs; then, an agent-based `RefineBot' updates them to remove shortcuts. Benchmarking on state-of-the-art MLLMs reveal a peak performance of 53\%; models with smaller LLMs only slightly underperform top models, suggesting that language-based reasoning is less challenging than multimodal reasoning; and tuning with scientific articles enhances performance. Expert analysis of chain-of-thought responses shows that perception errors are the most frequent, followed by knowledge errors and then overgeneralization errors. These insights highlight the challenges in multimodal scientific reasoning, showing MicroVQA is a valuable resource advancing AI-driven biomedical research. MicroVQA is available at https://huggingface.co/datasets/jmhb/microvqa, and project page at https://jmhb0.github.io/microvqa.
RadAdapt: Radiology Report Summarization via Lightweight Domain Adaptation of Large Language Models
May 02, 2023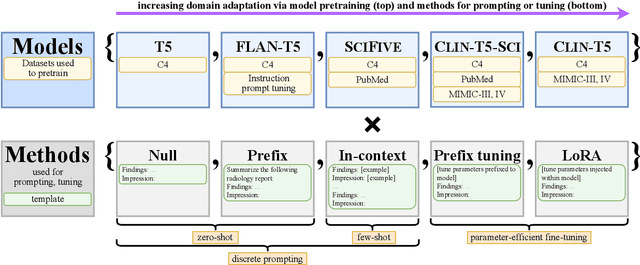
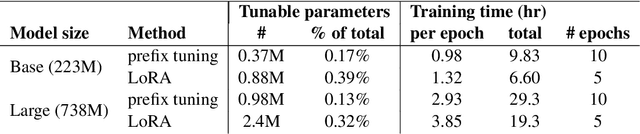
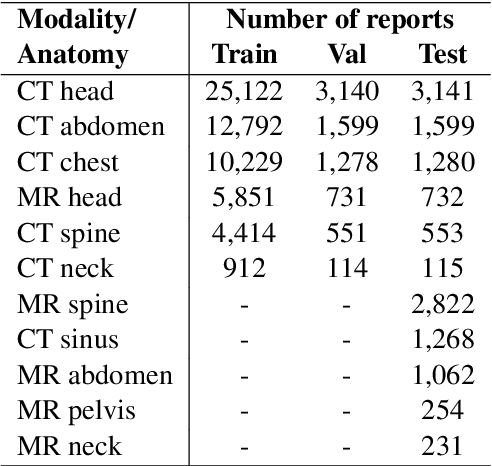

Abstract:We systematically investigate lightweight strategies to adapt large language models (LLMs) for the task of radiology report summarization (RRS). Specifically, we focus on domain adaptation via pretraining (on natural language, biomedical text, and clinical text) and via prompting (zero-shot, in-context learning) or parameter-efficient fine-tuning (prefix tuning, LoRA). Our results on the MIMIC-III dataset consistently demonstrate best performance by maximally adapting to the task via pretraining on clinical text and parameter-efficient fine-tuning on RRS examples. Importantly, this method fine-tunes a mere 0.32% of parameters throughout the model, in contrast to end-to-end fine-tuning (100% of parameters). Additionally, we study the effect of in-context examples and out-of-distribution (OOD) training before concluding with a radiologist reader study and qualitative analysis. Our findings highlight the importance of domain adaptation in RRS and provide valuable insights toward developing effective natural language processing solutions for clinical tasks.
DARCNN: Domain Adaptive Region-based Convolutional Neural Network for Unsupervised Instance Segmentation in Biomedical Images
Apr 03, 2021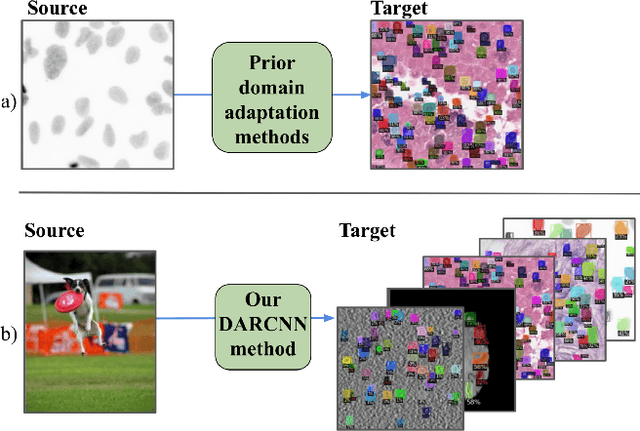
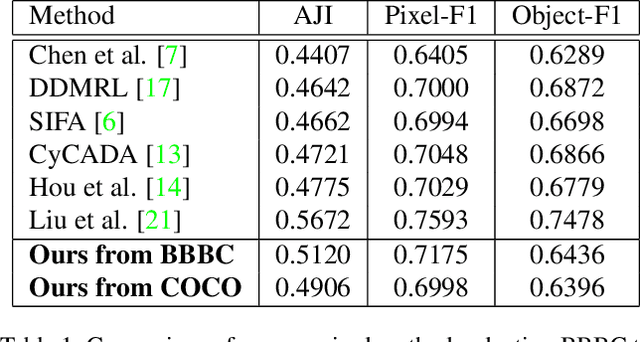
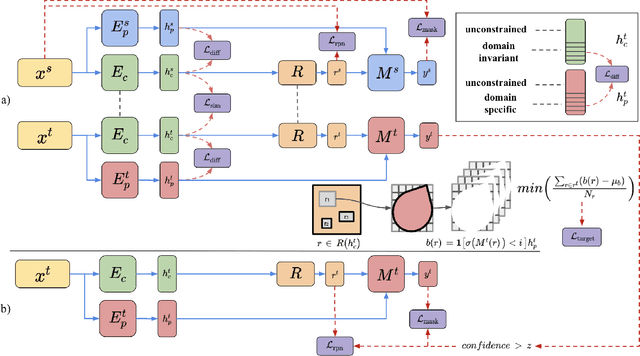
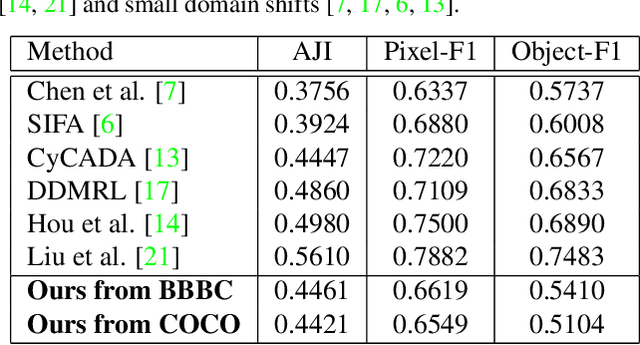
Abstract:In the biomedical domain, there is an abundance of dense, complex data where objects of interest may be challenging to detect or constrained by limits of human knowledge. Labelled domain specific datasets for supervised tasks are often expensive to obtain, and furthermore discovery of novel distinct objects may be desirable for unbiased scientific discovery. Therefore, we propose leveraging the wealth of annotations in benchmark computer vision datasets to conduct unsupervised instance segmentation for diverse biomedical datasets. The key obstacle is thus overcoming the large domain shift from common to biomedical images. We propose a Domain Adaptive Region-based Convolutional Neural Network (DARCNN), that adapts knowledge of object definition from COCO, a large labelled vision dataset, to multiple biomedical datasets. We introduce a domain separation module, a self-supervised representation consistency loss, and an augmented pseudo-labelling stage within DARCNN to effectively perform domain adaptation across such large domain shifts. We showcase DARCNN's performance for unsupervised instance segmentation on numerous biomedical datasets.
Learning Hyperbolic Representations for Unsupervised 3D Segmentation
Dec 04, 2020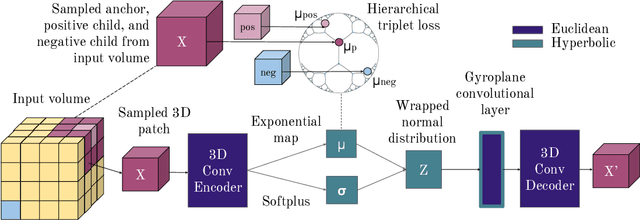
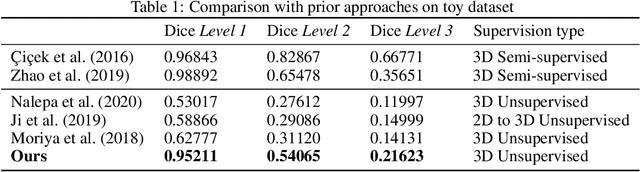
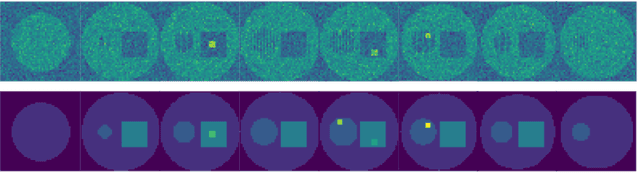
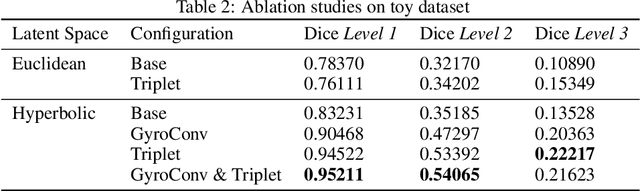
Abstract:There exists a need for unsupervised 3D segmentation on complex volumetric data, particularly when annotation ability is limited or discovery of new categories is desired. Using the observation that much of 3D volumetric data is innately hierarchical, we propose learning effective representations of 3D patches for unsupervised segmentation through a variational autoencoder (VAE) with a hyperbolic latent space and a proposed gyroplane convolutional layer, which better models the underlying hierarchical structure within a 3D image. We also introduce a hierarchical triplet loss and multi-scale patch sampling scheme to embed relationships across varying levels of granularity. We demonstrate the effectiveness of our hyperbolic representations for unsupervised 3D segmentation on a hierarchical toy dataset, BraTS whole tumor dataset, and cryogenic electron microscopy data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge