Viktoria Markova
DualTrack: Sensorless 3D Ultrasound needs Local and Global Context
Sep 11, 2025Abstract:Three-dimensional ultrasound (US) offers many clinical advantages over conventional 2D imaging, yet its widespread adoption is limited by the cost and complexity of traditional 3D systems. Sensorless 3D US, which uses deep learning to estimate a 3D probe trajectory from a sequence of 2D US images, is a promising alternative. Local features, such as speckle patterns, can help predict frame-to-frame motion, while global features, such as coarse shapes and anatomical structures, can situate the scan relative to anatomy and help predict its general shape. In prior approaches, global features are either ignored or tightly coupled with local feature extraction, restricting the ability to robustly model these two complementary aspects. We propose DualTrack, a novel dual-encoder architecture that leverages decoupled local and global encoders specialized for their respective scales of feature extraction. The local encoder uses dense spatiotemporal convolutions to capture fine-grained features, while the global encoder utilizes an image backbone (e.g., a 2D CNN or foundation model) and temporal attention layers to embed high-level anatomical features and long-range dependencies. A lightweight fusion module then combines these features to estimate the trajectory. Experimental results on a large public benchmark show that DualTrack achieves state-of-the-art accuracy and globally consistent 3D reconstructions, outperforming previous methods and yielding an average reconstruction error below 5 mm.
RIDE: Self-Supervised Learning of Rotation-Equivariant Keypoint Detection and Invariant Description for Endoscopy
Sep 18, 2023



Abstract:Unlike in natural images, in endoscopy there is no clear notion of an up-right camera orientation. Endoscopic videos therefore often contain large rotational motions, which require keypoint detection and description algorithms to be robust to these conditions. While most classical methods achieve rotation-equivariant detection and invariant description by design, many learning-based approaches learn to be robust only up to a certain degree. At the same time learning-based methods under moderate rotations often outperform classical approaches. In order to address this shortcoming, in this paper we propose RIDE, a learning-based method for rotation-equivariant detection and invariant description. Following recent advancements in group-equivariant learning, RIDE models rotation-equivariance implicitly within its architecture. Trained in a self-supervised manner on a large curation of endoscopic images, RIDE requires no manual labeling of training data. We test RIDE in the context of surgical tissue tracking on the SuPeR dataset as well as in the context of relative pose estimation on a repurposed version of the SCARED dataset. In addition we perform explicit studies showing its robustness to large rotations. Our comparison against recent learning-based and classical approaches shows that RIDE sets a new state-of-the-art performance on matching and relative pose estimation tasks and scores competitively on surgical tissue tracking.
Global Multi-modal 2D/3D Registration via Local Descriptors Learning
May 06, 2022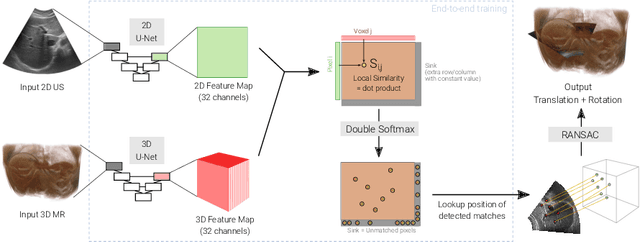
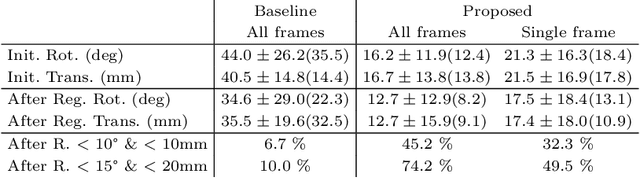
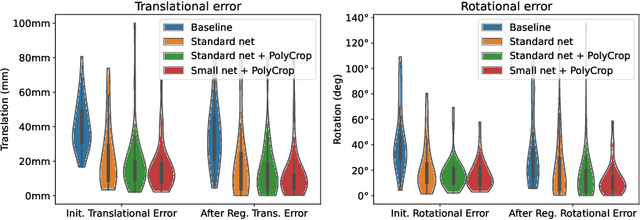
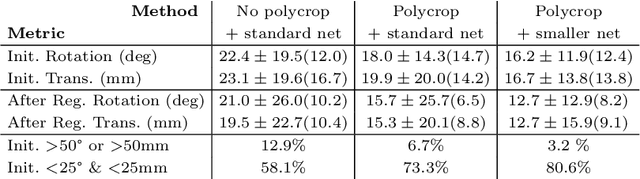
Abstract:Multi-modal registration is a required step for many image-guided procedures, especially ultrasound-guided interventions that require anatomical context. While a number of such registration algorithms are already available, they all require a good initialization to succeed due to the challenging appearance of ultrasound images and the arbitrary coordinate system they are acquired in. In this paper, we present a novel approach to solve the problem of registration of an ultrasound sweep to a pre-operative image. We learn dense keypoint descriptors from which we then estimate the registration. We show that our method overcomes the challenges inherent to registration tasks with freehand ultrasound sweeps, namely, the multi-modality and multidimensionality of the data in addition to lack of precise ground truth and low amounts of training examples. We derive a registration method that is fast, generic, fully automatic, does not require any initialization and can naturally generate visualizations aiding interpretability and explainability. Our approach is evaluated on a clinical dataset of paired MR volumes and ultrasound sequences.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge