Oliver Zettinig
DualTrack: Sensorless 3D Ultrasound needs Local and Global Context
Sep 11, 2025Abstract:Three-dimensional ultrasound (US) offers many clinical advantages over conventional 2D imaging, yet its widespread adoption is limited by the cost and complexity of traditional 3D systems. Sensorless 3D US, which uses deep learning to estimate a 3D probe trajectory from a sequence of 2D US images, is a promising alternative. Local features, such as speckle patterns, can help predict frame-to-frame motion, while global features, such as coarse shapes and anatomical structures, can situate the scan relative to anatomy and help predict its general shape. In prior approaches, global features are either ignored or tightly coupled with local feature extraction, restricting the ability to robustly model these two complementary aspects. We propose DualTrack, a novel dual-encoder architecture that leverages decoupled local and global encoders specialized for their respective scales of feature extraction. The local encoder uses dense spatiotemporal convolutions to capture fine-grained features, while the global encoder utilizes an image backbone (e.g., a 2D CNN or foundation model) and temporal attention layers to embed high-level anatomical features and long-range dependencies. A lightweight fusion module then combines these features to estimate the trajectory. Experimental results on a large public benchmark show that DualTrack achieves state-of-the-art accuracy and globally consistent 3D reconstructions, outperforming previous methods and yielding an average reconstruction error below 5 mm.
DISA: DIfferentiable Similarity Approximation for Universal Multimodal Registration
Jul 19, 2023



Abstract:Multimodal image registration is a challenging but essential step for numerous image-guided procedures. Most registration algorithms rely on the computation of complex, frequently non-differentiable similarity metrics to deal with the appearance discrepancy of anatomical structures between imaging modalities. Recent Machine Learning based approaches are limited to specific anatomy-modality combinations and do not generalize to new settings. We propose a generic framework for creating expressive cross-modal descriptors that enable fast deformable global registration. We achieve this by approximating existing metrics with a dot-product in the feature space of a small convolutional neural network (CNN) which is inherently differentiable can be trained without registered data. Our method is several orders of magnitude faster than local patch-based metrics and can be directly applied in clinical settings by replacing the similarity measure with the proposed one. Experiments on three different datasets demonstrate that our approach generalizes well beyond the training data, yielding a broad capture range even on unseen anatomies and modality pairs, without the need for specialized retraining. We make our training code and data publicly available.
PRO-TIP: Phantom for RObust automatic ultrasound calibration by TIP detection
Jun 13, 2022
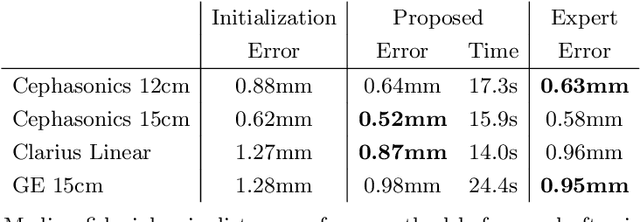
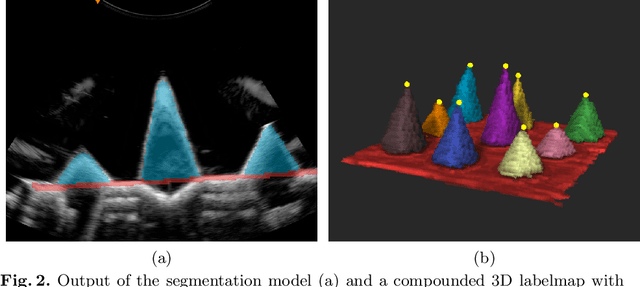
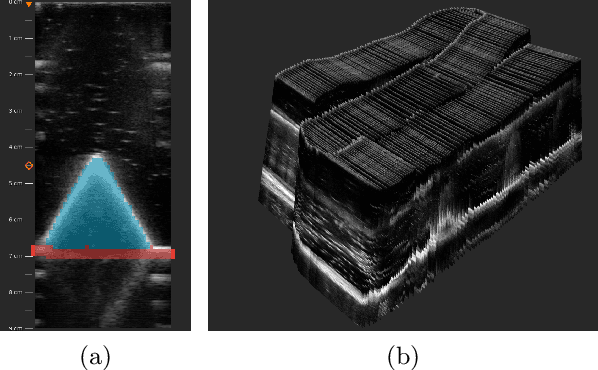
Abstract:We propose a novel method to automatically calibrate tracked ultrasound probes. To this end we design a custom phantom consisting of nine cones with different heights. The tips are used as key points to be matched between multiple sweeps. We extract them using a convolutional neural network to segment the cones in every ultrasound frame and then track them across the sweep. The calibration is robustly estimated using RANSAC and later refined employing image based techniques. Our phantom can be 3D-printed and offers many advantages over state-of-the-art methods. The phantom design and algorithm code are freely available online. Since our phantom does not require a tracking target on itself, ease of use is improved over currently used techniques. The fully automatic method generalizes to new probes and different vendors, as shown in our experiments. Our approach produces results comparable to calibrations obtained by a domain expert.
Global Multi-modal 2D/3D Registration via Local Descriptors Learning
May 06, 2022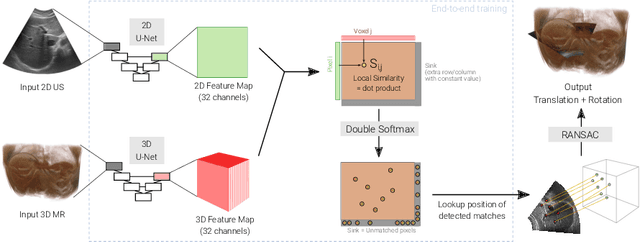
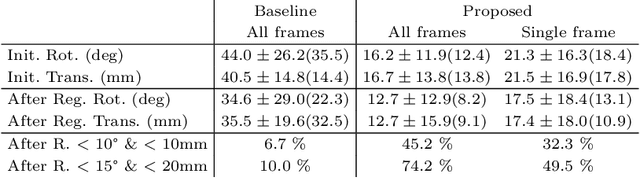
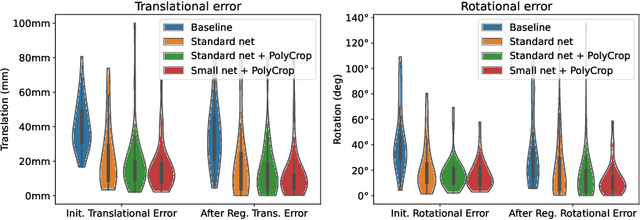
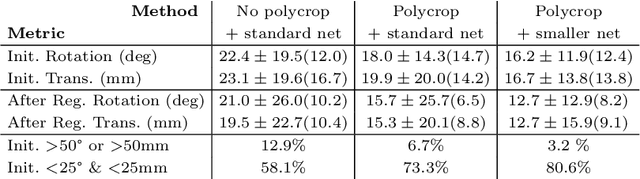
Abstract:Multi-modal registration is a required step for many image-guided procedures, especially ultrasound-guided interventions that require anatomical context. While a number of such registration algorithms are already available, they all require a good initialization to succeed due to the challenging appearance of ultrasound images and the arbitrary coordinate system they are acquired in. In this paper, we present a novel approach to solve the problem of registration of an ultrasound sweep to a pre-operative image. We learn dense keypoint descriptors from which we then estimate the registration. We show that our method overcomes the challenges inherent to registration tasks with freehand ultrasound sweeps, namely, the multi-modality and multidimensionality of the data in addition to lack of precise ground truth and low amounts of training examples. We derive a registration method that is fast, generic, fully automatic, does not require any initialization and can naturally generate visualizations aiding interpretability and explainability. Our approach is evaluated on a clinical dataset of paired MR volumes and ultrasound sequences.
Towards MRI-Based Autonomous Robotic US Acquisitions: A First Feasibility Study
Jul 28, 2016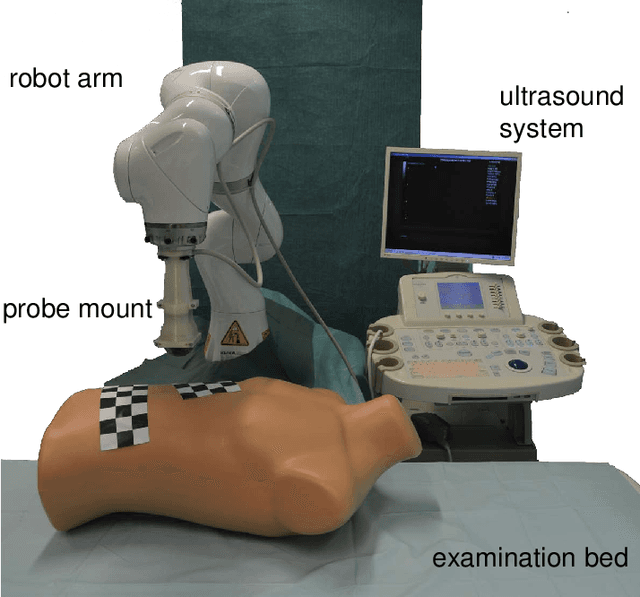
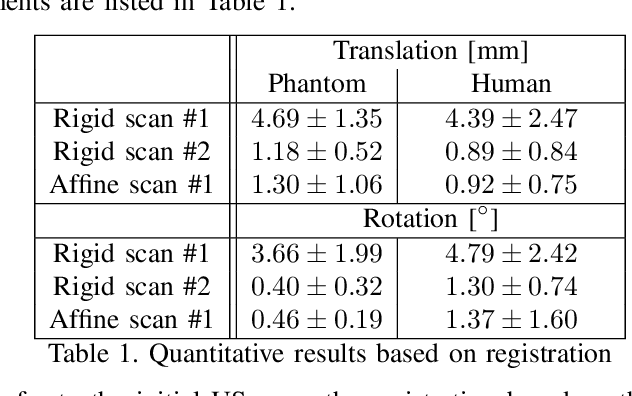
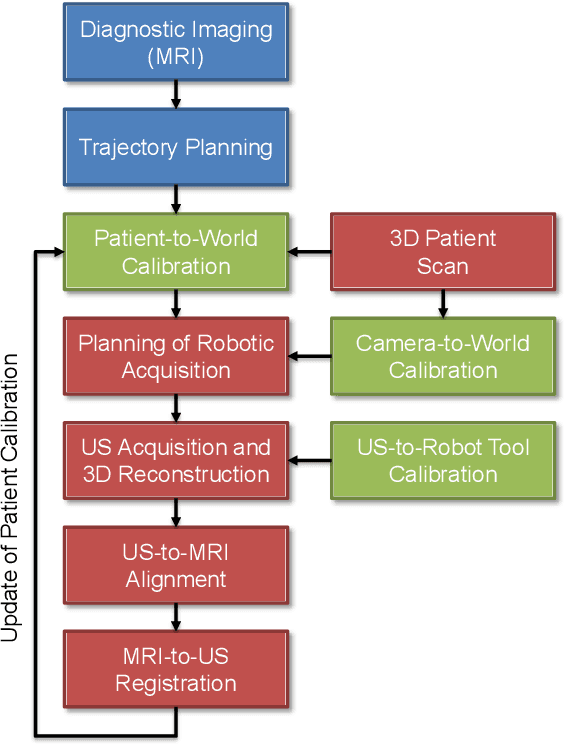
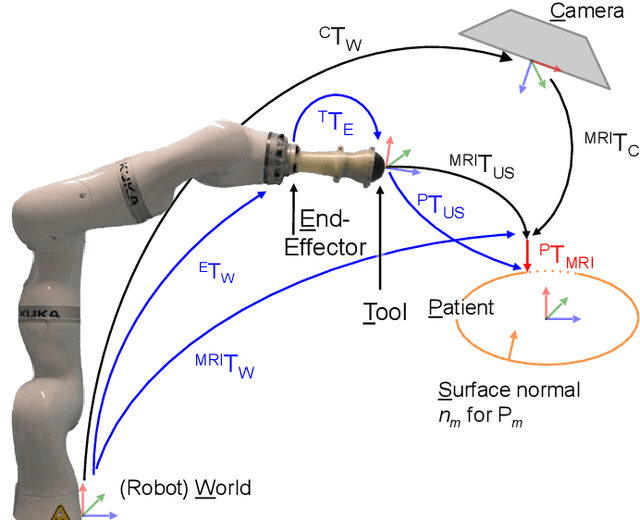
Abstract:Robotic ultrasound has the potential to assist and guide physicians during interventions. In this work, we present a set of methods and a workflow to enable autonomous MRI-guided ultrasound acquisitions. Our approach uses a structured-light 3D scanner for patient-to-robot and image-to-patient calibration, which in turn is used to plan 3D ultrasound trajectories. These MRI-based trajectories are followed autonomously by the robot and are further refined online using automatic MRI/US registration. Despite the low spatial resolution of structured light scanners, the initial planned acquisition path can be followed with an accuracy of 2.46 +/- 0.96 mm. This leads to a good initialization of the MRI/US registration: the 3D-scan-based alignment for planning and acquisition shows an accuracy (distance between planned ultrasound and MRI) of 4.47 mm, and 0.97 mm after an online-update of the calibration based on a closed loop registration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge