Massimo Filippi
Semi-Supervised Diffusion Model for Brain Age Prediction
Feb 14, 2024



Abstract:Brain age prediction models have succeeded in predicting clinical outcomes in neurodegenerative diseases, but can struggle with tasks involving faster progressing diseases and low quality data. To enhance their performance, we employ a semi-supervised diffusion model, obtaining a 0.83(p<0.01) correlation between chronological and predicted age on low quality T1w MR images. This was competitive with state-of-the-art non-generative methods. Furthermore, the predictions produced by our model were significantly associated with survival length (r=0.24, p<0.05) in Amyotrophic Lateral Sclerosis. Thus, our approach demonstrates the value of diffusion-based architectures for the task of brain age prediction.
Segmentation of Multiple Sclerosis Lesions across Hospitals: Learn Continually or Train from Scratch?
Oct 27, 2022



Abstract:Segmentation of Multiple Sclerosis (MS) lesions is a challenging problem. Several deep-learning-based methods have been proposed in recent years. However, most methods tend to be static, that is, a single model trained on a large, specialized dataset, which does not generalize well. Instead, the model should learn across datasets arriving sequentially from different hospitals by building upon the characteristics of lesions in a continual manner. In this regard, we explore experience replay, a well-known continual learning method, in the context of MS lesion segmentation across multi-contrast data from 8 different hospitals. Our experiments show that replay is able to achieve positive backward transfer and reduce catastrophic forgetting compared to sequential fine-tuning. Furthermore, replay outperforms the multi-domain training, thereby emerging as a promising solution for the segmentation of MS lesions. The code is available at this link: https://github.com/naga-karthik/continual-learning-ms
Multi-branch Convolutional Neural Network for Multiple Sclerosis Lesion Segmentation
Nov 16, 2018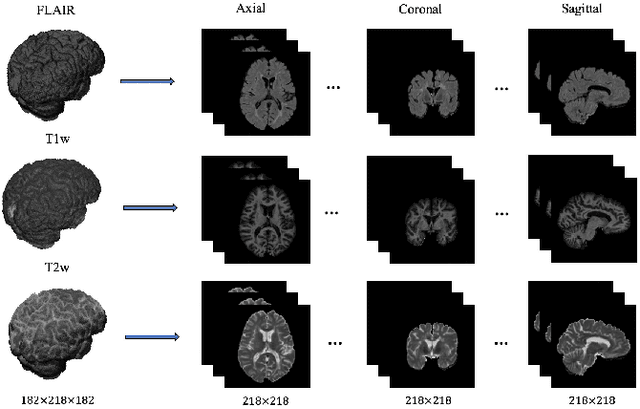
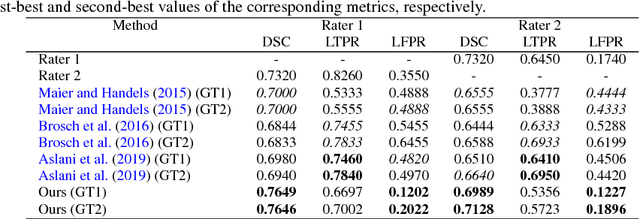
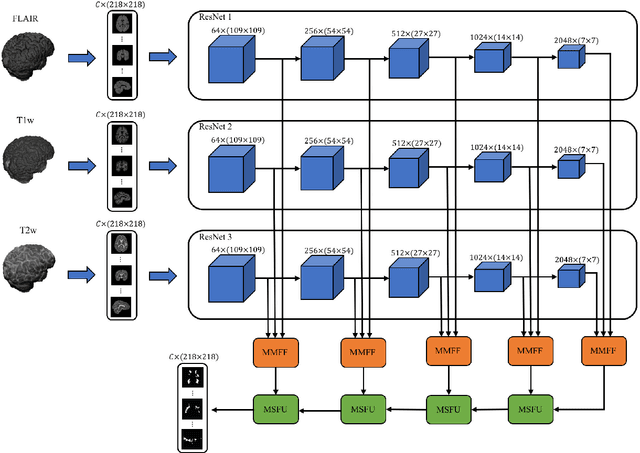
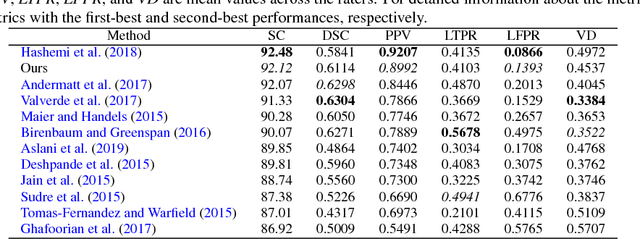
Abstract:In this paper, we present an automated approach for segmenting multiple sclerosis (MS) lesions from multi-modal brain magnetic resonance images. Our method is based on a deep end-to-end 2D convolutional neural network (CNN) for slice-based segmentation of 3D volumetric data. The proposed CNN includes a multi-branch downsampling path, which enables the network to encode slices from multiple modalities separately. Multi-scale feature fusion blocks are proposed to combine feature maps from different modalities at different stages of the network. Then, multi-scale feature upsampling blocks are introduced to upsize combined feature maps with different resolutions to leverage information from the lesion shape and location. We trained and tested our model using orthogonal plane orientations of each 3D modality to exploit the contextual information in all directions. The proposed pipeline is evaluated on two different datasets: a private dataset including 37 MS patients and a publicly available dataset known as the ISBI 2015 longitudinal MS lesion segmentation challenge dataset, consisting of 14 MS patients. Considering the ISBI challenge, at the time of submission, our method was amongst the top performing solutions. On the private dataset, using the same array of performance metrics as in the ISBI challenge, the proposed approach shows high improvements in MS lesion segmentation comparing with other publicly available tools.
Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks
Sep 11, 2018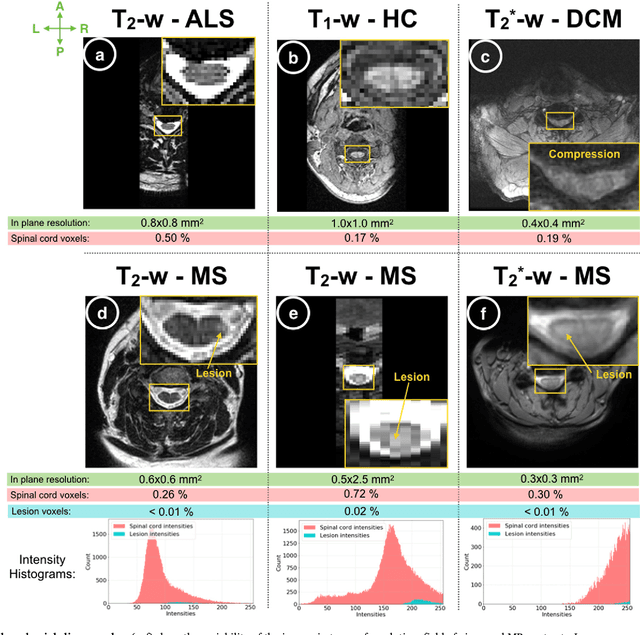
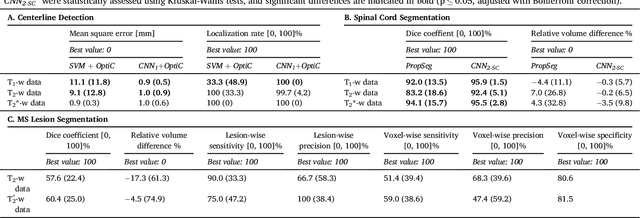
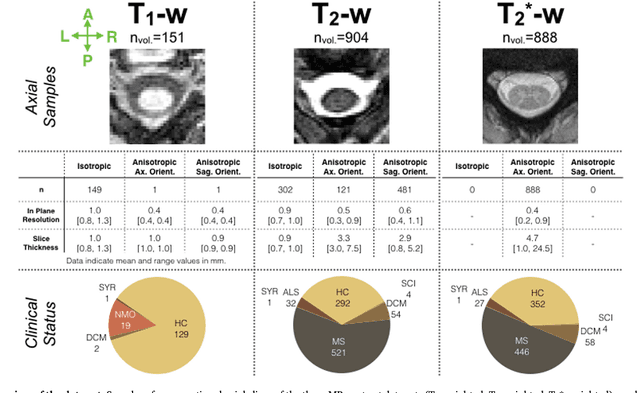
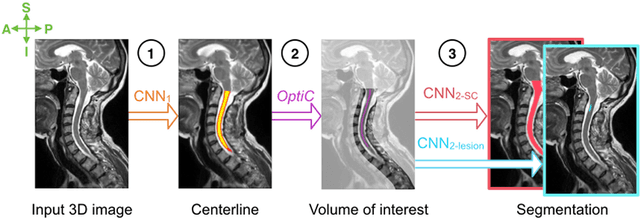
Abstract:The spinal cord is frequently affected by atrophy and/or lesions in multiple sclerosis (MS) patients. Segmentation of the spinal cord and lesions from MRI data provides measures of damage, which are key criteria for the diagnosis, prognosis, and longitudinal monitoring in MS. Automating this operation eliminates inter-rater variability and increases the efficiency of large-throughput analysis pipelines. Robust and reliable segmentation across multi-site spinal cord data is challenging because of the large variability related to acquisition parameters and image artifacts. The goal of this study was to develop a fully-automatic framework, robust to variability in both image parameters and clinical condition, for segmentation of the spinal cord and intramedullary MS lesions from conventional MRI data. Scans of 1,042 subjects (459 healthy controls, 471 MS patients, and 112 with other spinal pathologies) were included in this multi-site study (n=30). Data spanned three contrasts (T1-, T2-, and T2*-weighted) for a total of 1,943 volumes. The proposed cord and lesion automatic segmentation approach is based on a sequence of two Convolutional Neural Networks (CNNs). To deal with the very small proportion of spinal cord and/or lesion voxels compared to the rest of the volume, a first CNN with 2D dilated convolutions detects the spinal cord centerline, followed by a second CNN with 3D convolutions that segments the spinal cord and/or lesions. When compared against manual segmentation, our CNN-based approach showed a median Dice of 95% vs. 88% for PropSeg, a state-of-the-art spinal cord segmentation method. Regarding lesion segmentation on MS data, our framework provided a Dice of 60%, a relative volume difference of -15%, and a lesion-wise detection sensitivity and precision of 83% and 77%, respectively. The proposed framework is open-source and readily available in the Spinal Cord Toolbox.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge