Thomas R. Campion
Extracting Post-Acute Sequelae of SARS-CoV-2 Infection Symptoms from Clinical Notes via Hybrid Natural Language Processing
Aug 17, 2025Abstract:Accurately and efficiently diagnosing Post-Acute Sequelae of COVID-19 (PASC) remains challenging due to its myriad symptoms that evolve over long- and variable-time intervals. To address this issue, we developed a hybrid natural language processing pipeline that integrates rule-based named entity recognition with BERT-based assertion detection modules for PASC-symptom extraction and assertion detection from clinical notes. We developed a comprehensive PASC lexicon with clinical specialists. From 11 health systems of the RECOVER initiative network across the U.S., we curated 160 intake progress notes for model development and evaluation, and collected 47,654 progress notes for a population-level prevalence study. We achieved an average F1 score of 0.82 in one-site internal validation and 0.76 in 10-site external validation for assertion detection. Our pipeline processed each note at $2.448\pm 0.812$ seconds on average. Spearman correlation tests showed $\rho >0.83$ for positive mentions and $\rho >0.72$ for negative ones, both with $P <0.0001$. These demonstrate the effectiveness and efficiency of our models and their potential for improving PASC diagnosis.
DIRI: Adversarial Patient Reidentification with Large Language Models for Evaluating Clinical Text Anonymization
Oct 22, 2024Abstract:Sharing protected health information (PHI) is critical for furthering biomedical research. Before data can be distributed, practitioners often perform deidentification to remove any PHI contained in the text. Contemporary deidentification methods are evaluated on highly saturated datasets (tools achieve near-perfect accuracy) which may not reflect the full variability or complexity of real-world clinical text and annotating them is resource intensive, which is a barrier to real-world applications. To address this gap, we developed an adversarial approach using a large language model (LLM) to re-identify the patient corresponding to a redacted clinical note and evaluated the performance with a novel De-Identification/Re-Identification (DIRI) method. Our method uses a large language model to reidentify the patient corresponding to a redacted clinical note. We demonstrate our method on medical data from Weill Cornell Medicine anonymized with three deidentification tools: rule-based Philter and two deep-learning-based models, BiLSTM-CRF and ClinicalBERT. Although ClinicalBERT was the most effective, masking all identified PII, our tool still reidentified 9% of clinical notes Our study highlights significant weaknesses in current deidentification technologies while providing a tool for iterative development and improvement.
Evaluating the Portability of an NLP System for Processing Echocardiograms: A Retrospective, Multi-site Observational Study
Apr 02, 2019
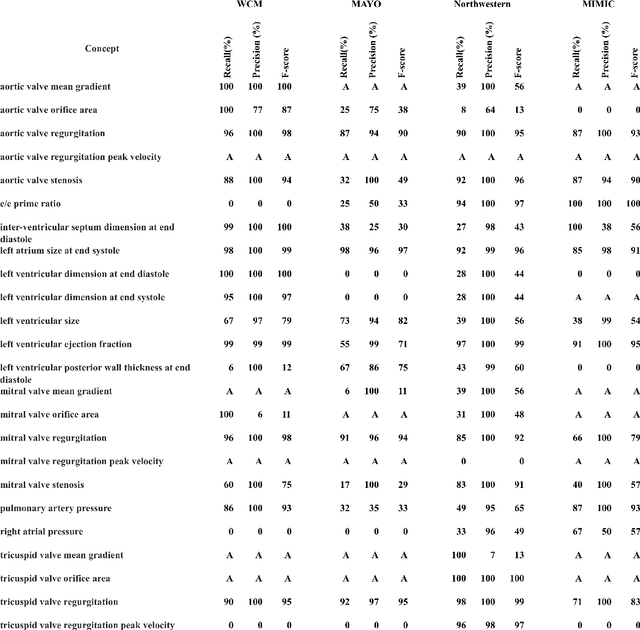
Abstract:While natural language processing (NLP) of unstructured clinical narratives holds the potential for patient care and clinical research, portability of NLP approaches across multiple sites remains a major challenge. This study investigated the portability of an NLP system developed initially at the Department of Veterans Affairs (VA) to extract 27 key cardiac concepts from free-text or semi-structured echocardiograms from three academic medical centers: Weill Cornell Medicine, Mayo Clinic and Northwestern Medicine. While the NLP system showed high precision and recall measurements for four target concepts (aortic valve regurgitation, left atrium size at end systole, mitral valve regurgitation, tricuspid valve regurgitation) across all sites, we found moderate or poor results for the remaining concepts and the NLP system performance varied between individual sites.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge