Tahsin M. Kurc
Evaluating histopathology transfer learning with ChampKit
Jun 14, 2022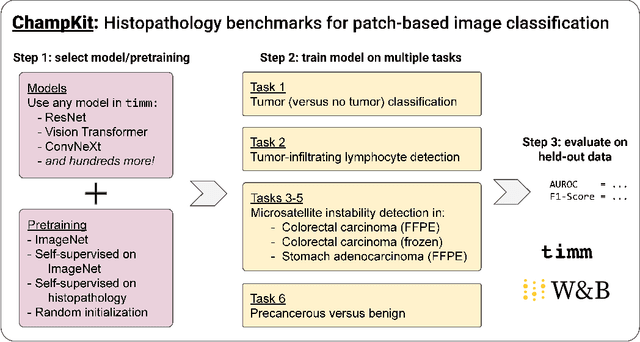
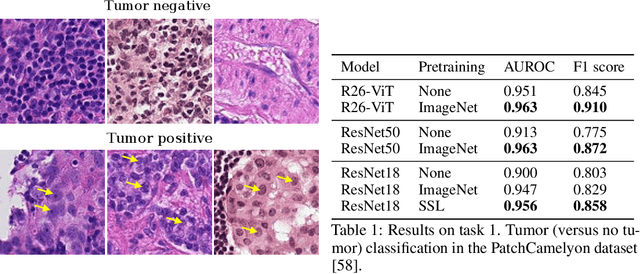
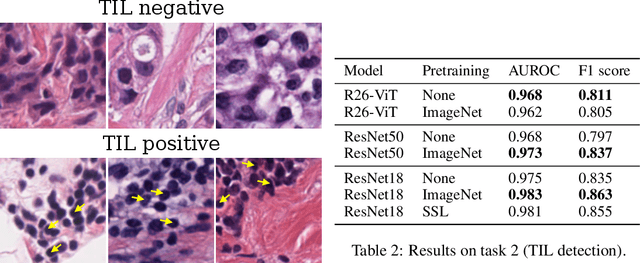
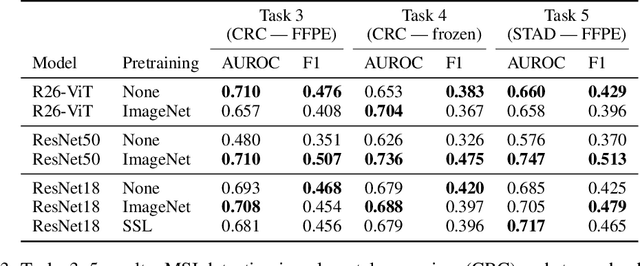
Abstract:Histopathology remains the gold standard for diagnosis of various cancers. Recent advances in computer vision, specifically deep learning, have facilitated the analysis of histopathology images for various tasks, including immune cell detection and microsatellite instability classification. The state-of-the-art for each task often employs base architectures that have been pretrained for image classification on ImageNet. The standard approach to develop classifiers in histopathology tends to focus narrowly on optimizing models for a single task, not considering the aspects of modeling innovations that improve generalization across tasks. Here we present ChampKit (Comprehensive Histopathology Assessment of Model Predictions toolKit): an extensible, fully reproducible benchmarking toolkit that consists of a broad collection of patch-level image classification tasks across different cancers. ChampKit enables a way to systematically document the performance impact of proposed improvements in models and methodology. ChampKit source code and data are freely accessible at https://github.com/kaczmarj/champkit .
Federated Learning for the Classification of Tumor Infiltrating Lymphocytes
Apr 01, 2022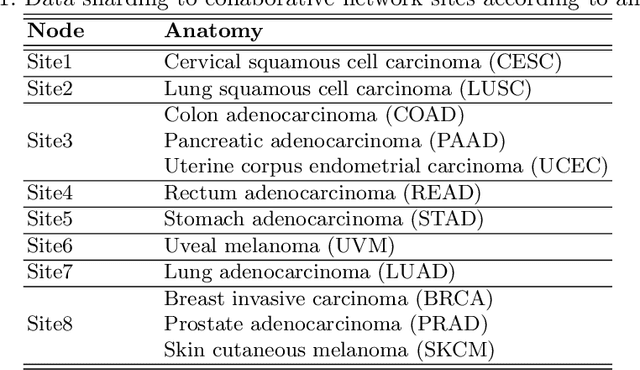
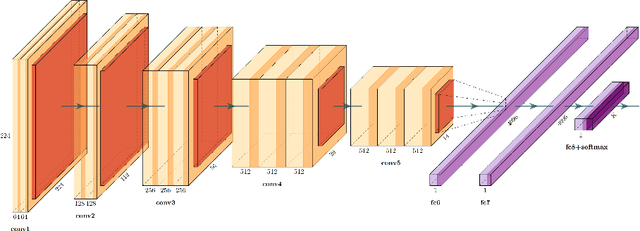
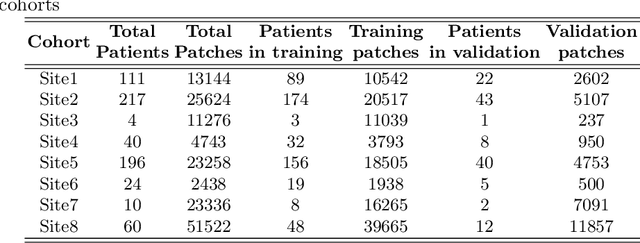

Abstract:We evaluate the performance of federated learning (FL) in developing deep learning models for analysis of digitized tissue sections. A classification application was considered as the example use case, on quantifiying the distribution of tumor infiltrating lymphocytes within whole slide images (WSIs). A deep learning classification model was trained using 50*50 square micron patches extracted from the WSIs. We simulated a FL environment in which a dataset, generated from WSIs of cancer from numerous anatomical sites available by The Cancer Genome Atlas repository, is partitioned in 8 different nodes. Our results show that the model trained with the federated training approach achieves similar performance, both quantitatively and qualitatively, to that of a model trained with all the training data pooled at a centralized location. Our study shows that FL has tremendous potential for enabling development of more robust and accurate models for histopathology image analysis without having to collect large and diverse training data at a single location.
Dataset of Segmented Nuclei in Hematoxylin and Eosin Stained Histopathology Images of 10 Cancer Types
Feb 18, 2020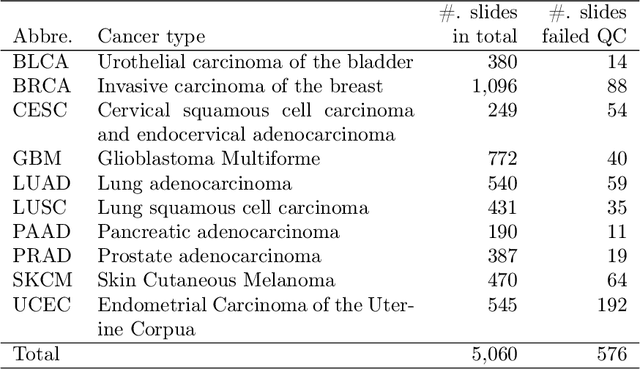
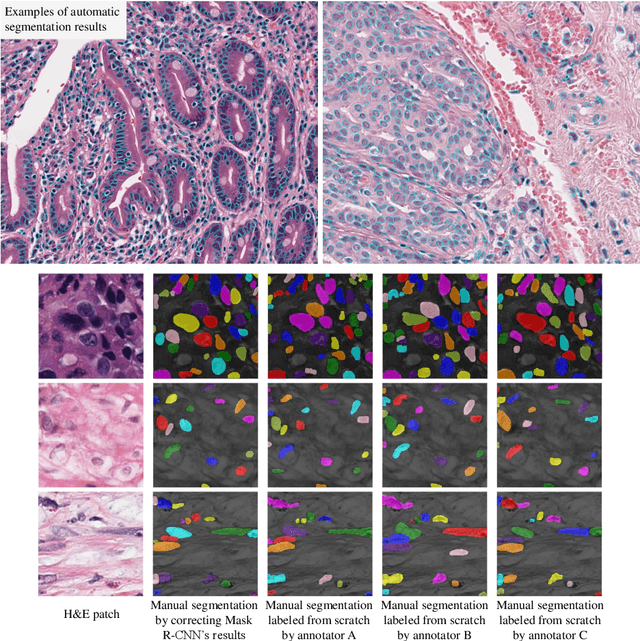
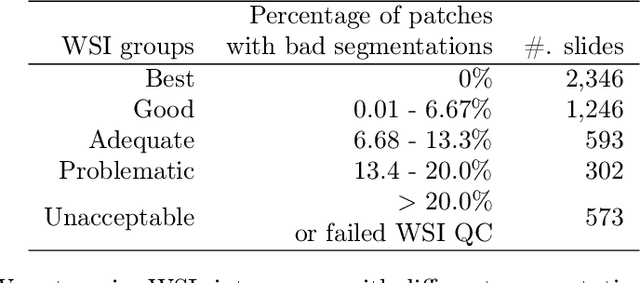
Abstract:The distribution and appearance of nuclei are essential markers for the diagnosis and study of cancer. Despite the importance of nuclear morphology, there is a lack of large scale, accurate, publicly accessible nucleus segmentation data. To address this, we developed an analysis pipeline that segments nuclei in whole slide tissue images from multiple cancer types with a quality control process. We have generated nucleus segmentation results in 5,060 Whole Slide Tissue images from 10 cancer types in The Cancer Genome Atlas. One key component of our work is that we carried out a multi-level quality control process (WSI-level and image patch-level), to evaluate the quality of our segmentation results. The image patch-level quality control used manual segmentation ground truth data from 1,356 sampled image patches. The datasets we publish in this work consist of roughly 5 billion quality controlled nuclei from more than 5,060 TCGA WSIs from 10 different TCGA cancer types and 1,356 manually segmented TCGA image patches from the same 10 cancer types plus additional 4 cancer types.
Unsupervised Histopathology Image Synthesis
Dec 13, 2017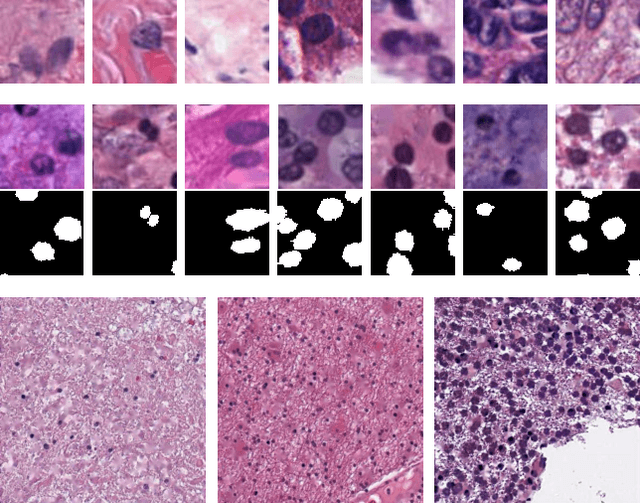
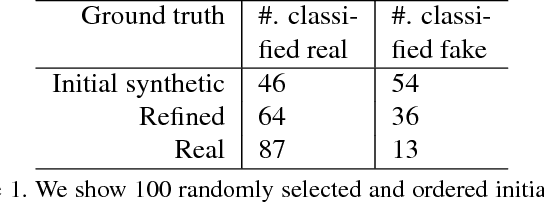
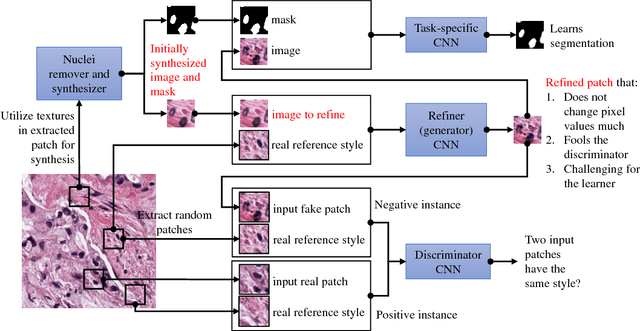
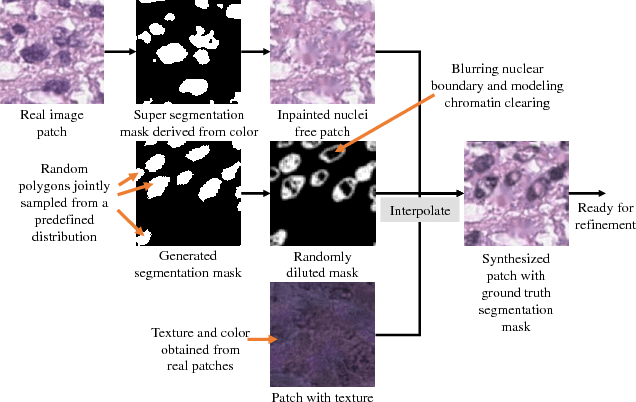
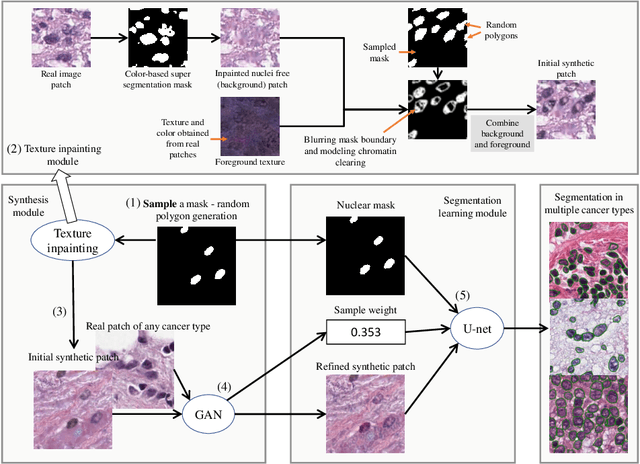
Abstract:Hematoxylin and Eosin stained histopathology image analysis is essential for the diagnosis and study of complicated diseases such as cancer. Existing state-of-the-art approaches demand extensive amount of supervised training data from trained pathologists. In this work we synthesize in an unsupervised manner, large histopathology image datasets, suitable for supervised training tasks. We propose a unified pipeline that: a) generates a set of initial synthetic histopathology images with paired information about the nuclei such as segmentation masks; b) refines the initial synthetic images through a Generative Adversarial Network (GAN) to reference styles; c) trains a task-specific CNN and boosts the performance of the task-specific CNN with on-the-fly generated adversarial examples. Our main contribution is that the synthetic images are not only realistic, but also representative (in reference styles) and relatively challenging for training task-specific CNNs. We test our method for nucleus segmentation using images from four cancer types. When no supervised data exists for a cancer type, our method without supervision cost significantly outperforms supervised methods which perform across-cancer generalization. Even when supervised data exists for all cancer types, our approach without supervision cost performs better than supervised methods.
Sparse Autoencoder for Unsupervised Nucleus Detection and Representation in Histopathology Images
Apr 10, 2017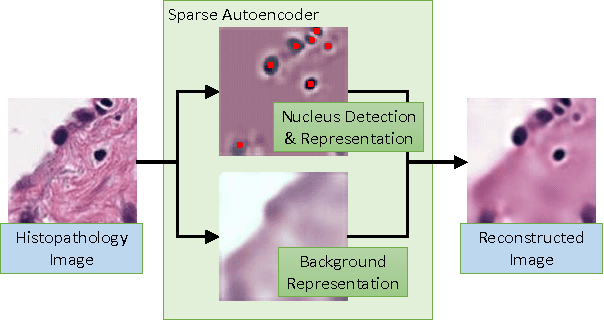
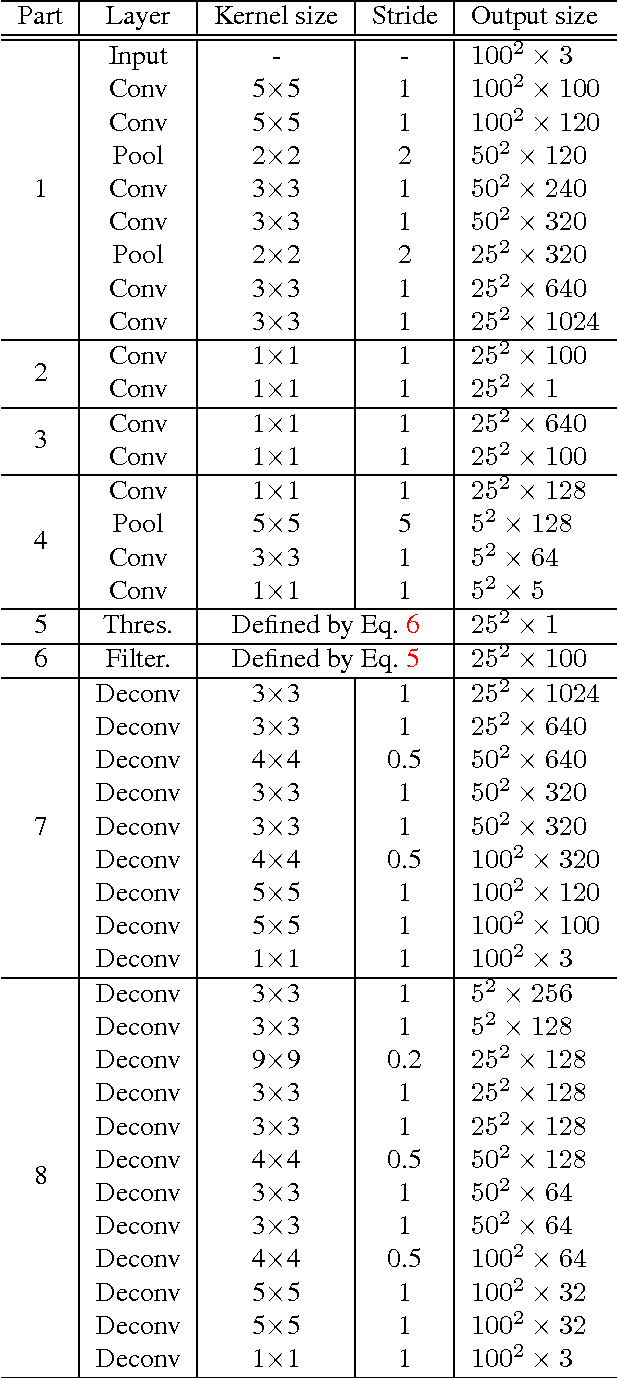
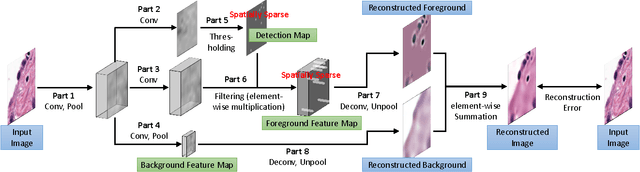
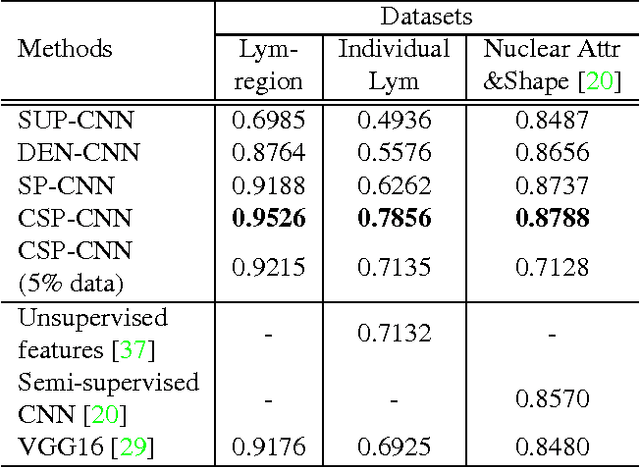
Abstract:Histopathology images are crucial to the study of complex diseases such as cancer. The histologic characteristics of nuclei play a key role in disease diagnosis, prognosis and analysis. In this work, we propose a sparse Convolutional Autoencoder (CAE) for fully unsupervised, simultaneous nucleus detection and feature extraction in histopathology tissue images. Our CAE detects and encodes nuclei in image patches in tissue images into sparse feature maps that encode both the location and appearance of nuclei. Our CAE is the first unsupervised detection network for computer vision applications. The pretrained nucleus detection and feature extraction modules in our CAE can be fine-tuned for supervised learning in an end-to-end fashion. We evaluate our method on four datasets and reduce the errors of state-of-the-art methods up to 42%. We are able to achieve comparable performance with only 5% of the fully-supervised annotation cost.
Center-Focusing Multi-task CNN with Injected Features for Classification of Glioma Nuclear Images
Jan 10, 2017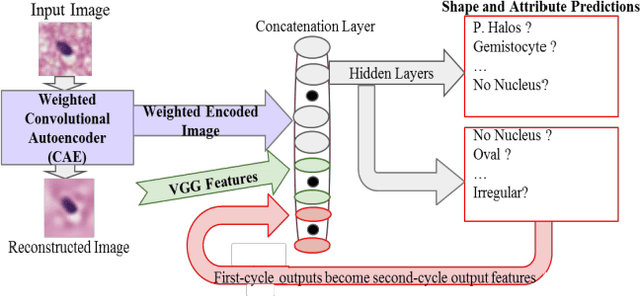
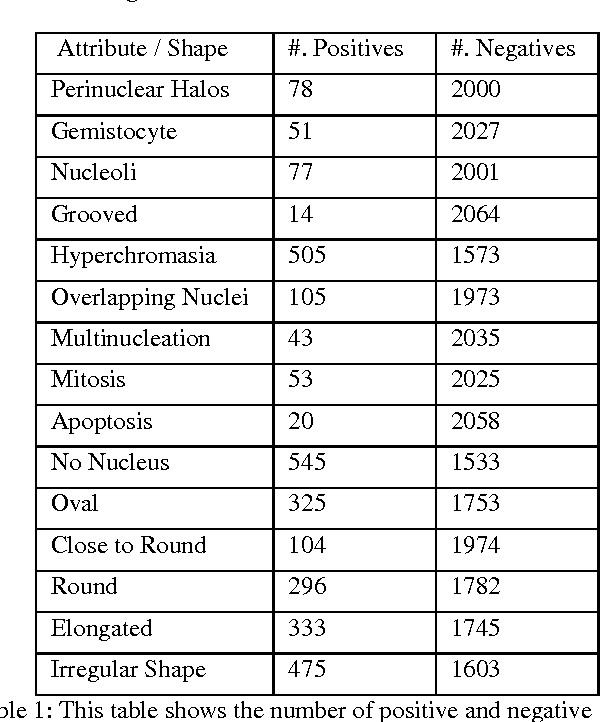
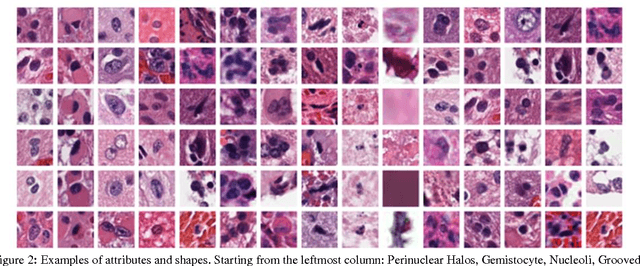
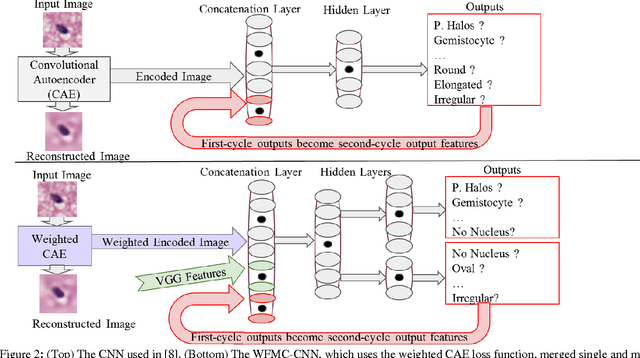
Abstract:Classifying the various shapes and attributes of a glioma cell nucleus is crucial for diagnosis and understanding the disease. We investigate automated classification of glioma nuclear shapes and visual attributes using Convolutional Neural Networks (CNNs) on pathology images of automatically segmented nuclei. We propose three methods that improve the performance of a previously-developed semi-supervised CNN. First, we propose a method that allows the CNN to focus on the most important part of an image- the image's center containing the nucleus. Second, we inject (concatenate) pre-extracted VGG features into an intermediate layer of our Semi-Supervised CNN so that during training, the CNN can learn a set of complementary features. Third, we separate the losses of the two groups of target classes (nuclear shapes and attributes) into a single-label loss and a multi-label loss so that the prior knowledge of inter-label exclusiveness can be incorporated. On a dataset of 2078 images, the proposed methods combined reduce the error rate of attribute and shape classification by 21.54% and 15.07% respectively compared to the existing state-of-the-art method on the same dataset.
Neural Networks with Smooth Adaptive Activation Functions for Regression
Aug 23, 2016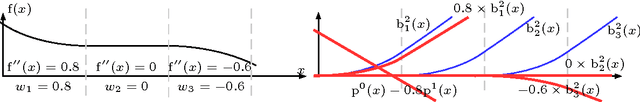
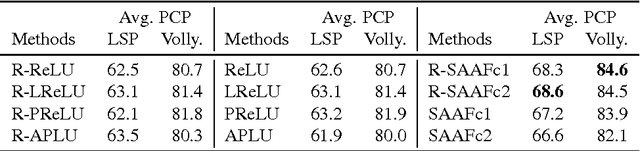
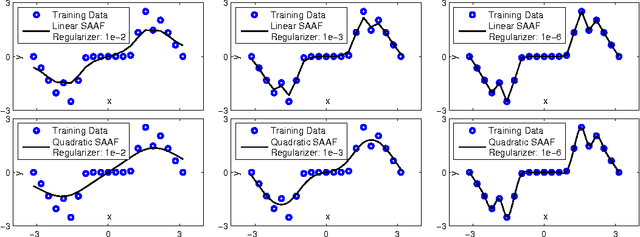
Abstract:In Neural Networks (NN), Adaptive Activation Functions (AAF) have parameters that control the shapes of activation functions. These parameters are trained along with other parameters in the NN. AAFs have improved performance of Neural Networks (NN) in multiple classification tasks. In this paper, we propose and apply AAFs on feedforward NNs for regression tasks. We argue that applying AAFs in the regression (second-to-last) layer of a NN can significantly decrease the bias of the regression NN. However, using existing AAFs may lead to overfitting. To address this problem, we propose a Smooth Adaptive Activation Function (SAAF) with piecewise polynomial form which can approximate any continuous function to arbitrary degree of error. NNs with SAAFs can avoid overfitting by simply regularizing the parameters. In particular, an NN with SAAFs is Lipschitz continuous given a bounded magnitude of the NN parameters. We prove an upper-bound for model complexity in terms of fat-shattering dimension for any Lipschitz continuous regression model. Thus, regularizing the parameters in NNs with SAAFs avoids overfitting. We empirically evaluated NNs with SAAFs and achieved state-of-the-art results on multiple regression datasets.
Patch-based Convolutional Neural Network for Whole Slide Tissue Image Classification
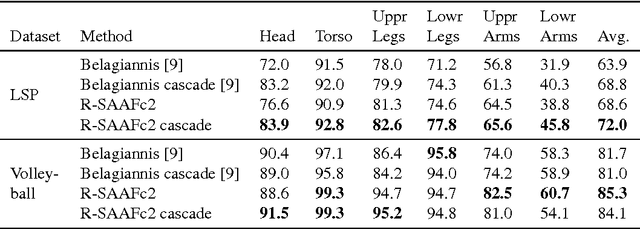
Mar 09, 2016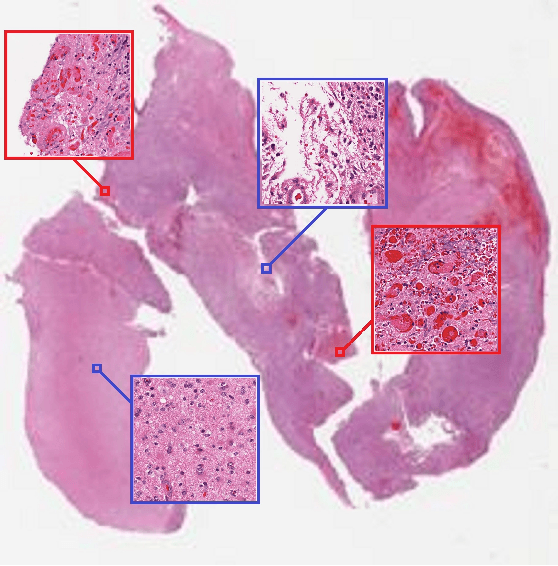


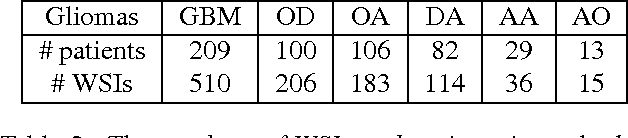
Abstract:Convolutional Neural Networks (CNN) are state-of-the-art models for many image classification tasks. However, to recognize cancer subtypes automatically, training a CNN on gigapixel resolution Whole Slide Tissue Images (WSI) is currently computationally impossible. The differentiation of cancer subtypes is based on cellular-level visual features observed on image patch scale. Therefore, we argue that in this situation, training a patch-level classifier on image patches will perform better than or similar to an image-level classifier. The challenge becomes how to intelligently combine patch-level classification results and model the fact that not all patches will be discriminative. We propose to train a decision fusion model to aggregate patch-level predictions given by patch-level CNNs, which to the best of our knowledge has not been shown before. Furthermore, we formulate a novel Expectation-Maximization (EM) based method that automatically locates discriminative patches robustly by utilizing the spatial relationships of patches. We apply our method to the classification of glioma and non-small-cell lung carcinoma cases into subtypes. The classification accuracy of our method is similar to the inter-observer agreement between pathologists. Although it is impossible to train CNNs on WSIs, we experimentally demonstrate using a comparable non-cancer dataset of smaller images that a patch-based CNN can outperform an image-based CNN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge