Stefan Klus
Bayesian Transfer Operators in Reproducing Kernel Hilbert Spaces
Sep 26, 2025Abstract:The Koopman operator, as a linear representation of a nonlinear dynamical system, has been attracting attention in many fields of science. Recently, Koopman operator theory has been combined with another concept that is popular in data science: reproducing kernel Hilbert spaces. We follow this thread into Gaussian process methods, and illustrate how these methods can alleviate two pervasive problems with kernel-based Koopman algorithms. The first being sparsity: most kernel methods do not scale well and require an approximation to become practical. We show that not only can the computational demands be reduced, but also demonstrate improved resilience against sensor noise. The second problem involves hyperparameter optimization and dictionary learning to adapt the model to the dynamical system. In summary, the main contribution of this work is the unification of Gaussian process regression and dynamic mode decomposition.
Data-driven system identification using quadratic embeddings of nonlinear dynamics
Jan 14, 2025



Abstract:We propose a novel data-driven method called QENDy (Quadratic Embedding of Nonlinear Dynamics) that not only allows us to learn quadratic representations of highly nonlinear dynamical systems, but also to identify the governing equations. The approach is based on an embedding of the system into a higher-dimensional feature space in which the dynamics become quadratic. Just like SINDy (Sparse Identification of Nonlinear Dynamics), our method requires trajectory data, time derivatives for the training data points, which can also be estimated using finite difference approximations, and a set of preselected basis functions, called dictionary. We illustrate the efficacy and accuracy of QENDy with the aid of various benchmark problems and compare its performance with SINDy and a deep learning method for identifying quadratic embeddings. Furthermore, we analyze the convergence of QENDy and SINDy in the infinite data limit, highlight their similarities and main differences, and compare the quadratic embedding with linearization techniques based on the Koopman operator.
Learning dynamical systems from data: Gradient-based dictionary optimization
Nov 07, 2024



Abstract:The Koopman operator plays a crucial role in analyzing the global behavior of dynamical systems. Existing data-driven methods for approximating the Koopman operator or discovering the governing equations of the underlying system typically require a fixed set of basis functions, also called dictionary. The optimal choice of basis functions is highly problem-dependent and often requires domain knowledge. We present a novel gradient descent-based optimization framework for learning suitable and interpretable basis functions from data and show how it can be used in combination with EDMD, SINDy, and PDE-FIND. We illustrate the efficacy of the proposed approach with the aid of various benchmark problems such as the Ornstein-Uhlenbeck process, Chua's circuit, a nonlinear heat equation, as well as protein-folding data.
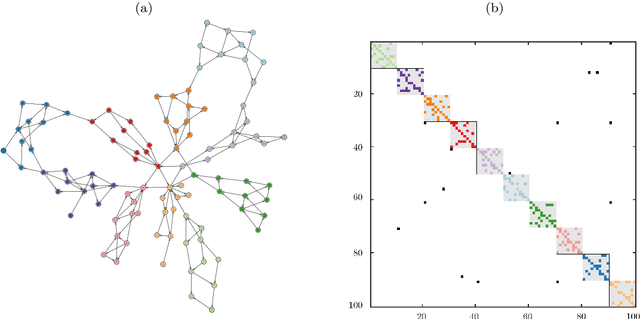
Clustering Time-Evolving Networks Using the Dynamic Graph Laplacian
Jul 12, 2024Abstract:Time-evolving graphs arise frequently when modeling complex dynamical systems such as social networks, traffic flow, and biological processes. Developing techniques to identify and analyze communities in these time-varying graph structures is an important challenge. In this work, we generalize existing spectral clustering algorithms from static to dynamic graphs using canonical correlation analysis (CCA) to capture the temporal evolution of clusters. Based on this extended canonical correlation framework, we define the dynamic graph Laplacian and investigate its spectral properties. We connect these concepts to dynamical systems theory via transfer operators, and illustrate the advantages of our method on benchmark graphs by comparison with existing methods. We show that the dynamic graph Laplacian allows for a clear interpretation of cluster structure evolution over time for directed and undirected graphs.
Dynamical systems and complex networks: A Koopman operator perspective
May 14, 2024Abstract:The Koopman operator has entered and transformed many research areas over the last years. Although the underlying concept$\unicode{x2013}$representing highly nonlinear dynamical systems by infinite-dimensional linear operators$\unicode{x2013}$has been known for a long time, the availability of large data sets and efficient machine learning algorithms for estimating the Koopman operator from data make this framework extremely powerful and popular. Koopman operator theory allows us to gain insights into the characteristic global properties of a system without requiring detailed mathematical models. We will show how these methods can also be used to analyze complex networks and highlight relationships between Koopman operators and graph Laplacians.
Transfer operators on graphs: Spectral clustering and beyond
May 19, 2023Abstract:Graphs and networks play an important role in modeling and analyzing complex interconnected systems such as transportation networks, integrated circuits, power grids, citation graphs, and biological and artificial neural networks. Graph clustering algorithms can be used to detect groups of strongly connected vertices and to derive coarse-grained models. We define transfer operators such as the Koopman operator and the Perron-Frobenius operator on graphs, study their spectral properties, introduce Galerkin projections of these operators, and illustrate how reduced representations can be estimated from data. In particular, we show that spectral clustering of undirected graphs can be interpreted in terms of eigenfunctions of the Koopman operator and propose novel clustering algorithms for directed graphs based on generalized transfer operators. We demonstrate the efficacy of the resulting algorithms on several benchmark problems and provide different interpretations of clusters.
Koopman-based spectral clustering of directed and time-evolving graphs
Apr 06, 2022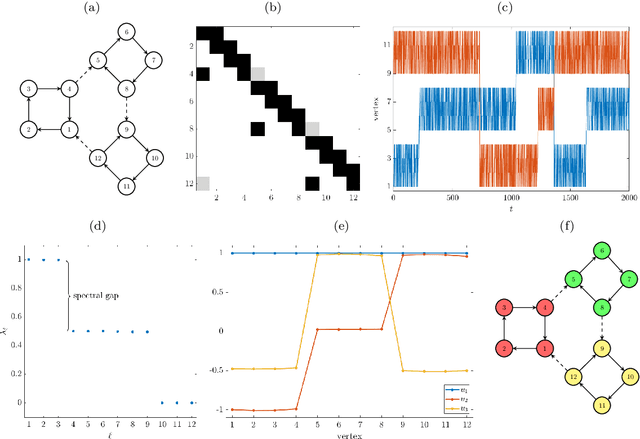
Abstract:While spectral clustering algorithms for undirected graphs are well established and have been successfully applied to unsupervised machine learning problems ranging from image segmentation and genome sequencing to signal processing and social network analysis, clustering directed graphs remains notoriously difficult. Two of the main challenges are that the eigenvalues and eigenvectors of graph Laplacians associated with directed graphs are in general complex-valued and that there is no universally accepted definition of clusters in directed graphs. We first exploit relationships between the graph Laplacian and transfer operators and in particular between clusters in undirected graphs and metastable sets in stochastic dynamical systems and then use a generalization of the notion of metastability to derive clustering algorithms for directed and time-evolving graphs. The resulting clusters can be interpreted as coherent sets, which play an important role in the analysis of transport and mixing processes in fluid flows.
A Dynamic Mode Decomposition Approach for Decentralized Spectral Clustering of Graphs
Feb 26, 2022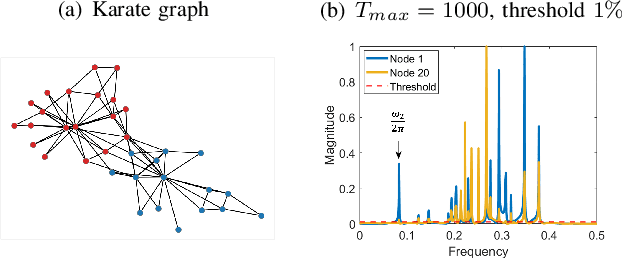
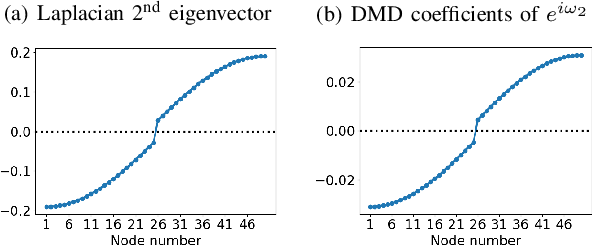
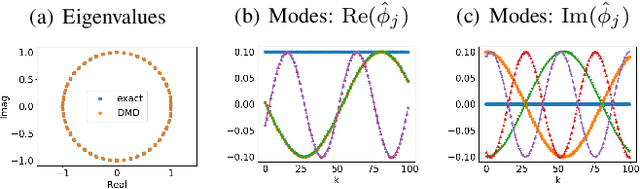
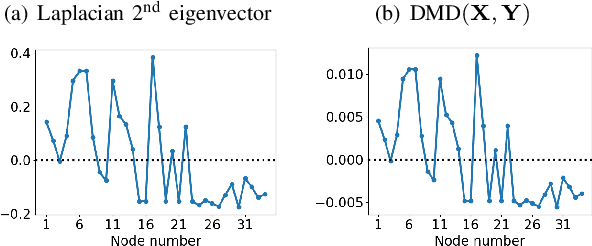
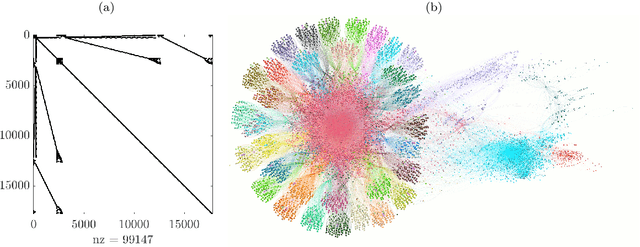
Abstract:We propose a novel robust decentralized graph clustering algorithm that is provably equivalent to the popular spectral clustering approach. Our proposed method uses the existing wave equation clustering algorithm that is based on propagating waves through the graph. However, instead of using a fast Fourier transform (FFT) computation at every node, our proposed approach exploits the Koopman operator framework. Specifically, we show that propagating waves in the graph followed by a local dynamic mode decomposition (DMD) computation at every node is capable of retrieving the eigenvalues and the local eigenvector components of the graph Laplacian, thereby providing local cluster assignments for all nodes. We demonstrate that the DMD computation is more robust than the existing FFT based approach and requires 20 times fewer steps of the wave equation to accurately recover the clustering information and reduces the relative error by orders of magnitude. We demonstrate the decentralized approach on a range of graph clustering problems.
Deeptime: a Python library for machine learning dynamical models from time series data
Oct 28, 2021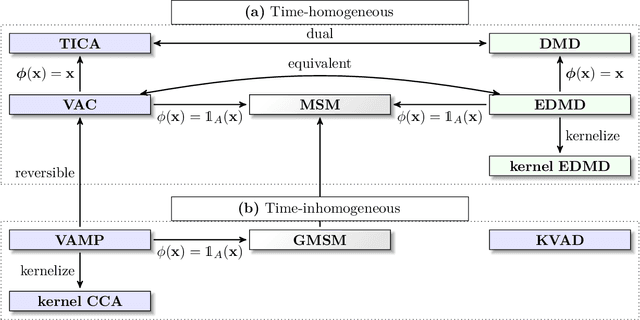

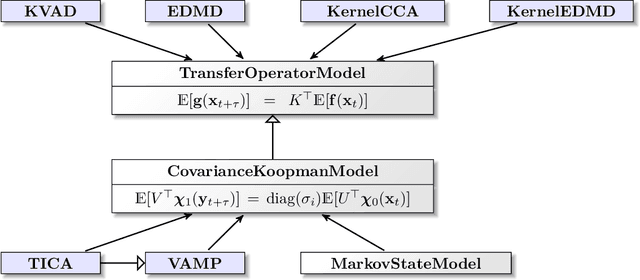
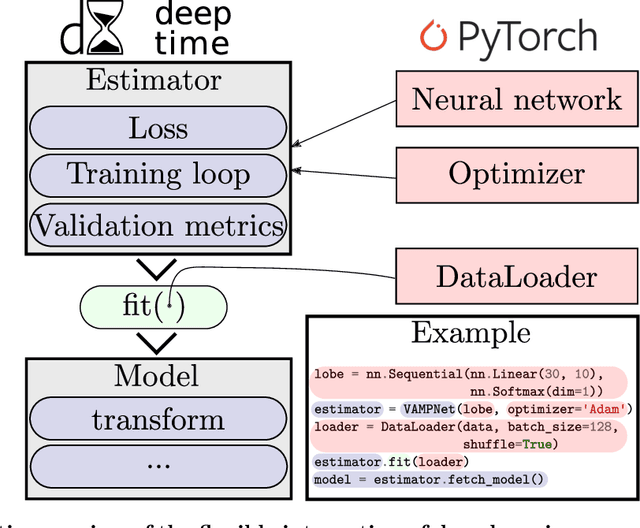
Abstract:Generation and analysis of time-series data is relevant to many quantitative fields ranging from economics to fluid mechanics. In the physical sciences, structures such as metastable and coherent sets, slow relaxation processes, collective variables dominant transition pathways or manifolds and channels of probability flow can be of great importance for understanding and characterizing the kinetic, thermodynamic and mechanistic properties of the system. Deeptime is a general purpose Python library offering various tools to estimate dynamical models based on time-series data including conventional linear learning methods, such as Markov state models (MSMs), Hidden Markov Models and Koopman models, as well as kernel and deep learning approaches such as VAMPnets and deep MSMs. The library is largely compatible with scikit-learn, having a range of Estimator classes for these different models, but in contrast to scikit-learn also provides deep Model classes, e.g. in the case of an MSM, which provide a multitude of analysis methods to compute interesting thermodynamic, kinetic and dynamical quantities, such as free energies, relaxation times and transition paths. The library is designed for ease of use but also easily maintainable and extensible code. In this paper we introduce the main features and structure of the deeptime software.
Symmetric and antisymmetric kernels for machine learning problems in quantum physics and chemistry
Mar 31, 2021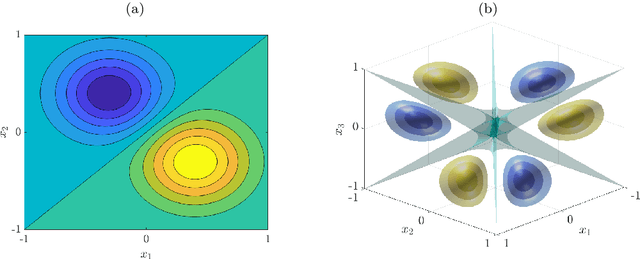
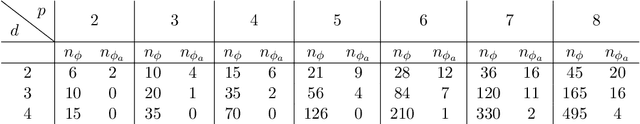
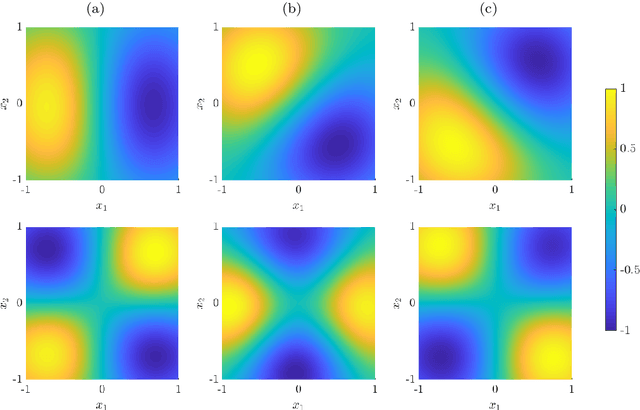
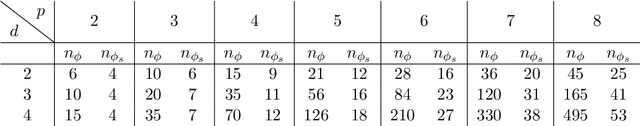
Abstract:We derive symmetric and antisymmetric kernels by symmetrizing and antisymmetrizing conventional kernels and analyze their properties. In particular, we compute the feature space dimensions of the resulting polynomial kernels, prove that the reproducing kernel Hilbert spaces induced by symmetric and antisymmetric Gaussian kernels are dense in the space of symmetric and antisymmetric functions, and propose a Slater determinant representation of the antisymmetric Gaussian kernel, which allows for an efficient evaluation even if the state space is high-dimensional. Furthermore, we show that by exploiting symmetries or antisymmetries the size of the training data set can be significantly reduced. The results are illustrated with guiding examples and simple quantum physics and chemistry applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge