Frank Noé
Enhanced Diffusion Sampling: Efficient Rare Event Sampling and Free Energy Calculation with Diffusion Models
Feb 18, 2026Abstract:The rare-event sampling problem has long been the central limiting factor in molecular dynamics (MD), especially in biomolecular simulation. Recently, diffusion models such as BioEmu have emerged as powerful equilibrium samplers that generate independent samples from complex molecular distributions, eliminating the cost of sampling rare transition events. However, a sampling problem remains when computing observables that rely on states which are rare in equilibrium, for example folding free energies. Here, we introduce enhanced diffusion sampling, enabling efficient exploration of rare-event regions while preserving unbiased thermodynamic estimators. The key idea is to perform quantitatively accurate steering protocols to generate biased ensembles and subsequently recover equilibrium statistics via exact reweighting. We instantiate our framework in three algorithms: UmbrellaDiff (umbrella sampling with diffusion models), $Δ$G-Diff (free-energy differences via tilted ensembles), and MetaDiff (a batchwise analogue for metadynamics). Across toy systems, protein folding landscapes and folding free energies, our methods achieve fast, accurate, and scalable estimation of equilibrium properties within GPU-minutes to hours per system -- closing the rare-event sampling gap that remained after the advent of diffusion-model equilibrium samplers.
Learning Hamiltonian Flow Maps: Mean Flow Consistency for Large-Timestep Molecular Dynamics
Jan 29, 2026Abstract:Simulating the long-time evolution of Hamiltonian systems is limited by the small timesteps required for stable numerical integration. To overcome this constraint, we introduce a framework to learn Hamiltonian Flow Maps by predicting the mean phase-space evolution over a chosen time span $Δt$, enabling stable large-timestep updates far beyond the stability limits of classical integrators. To this end, we impose a Mean Flow consistency condition for time-averaged Hamiltonian dynamics. Unlike prior approaches, this allows training on independent phase-space samples without access to future states, avoiding expensive trajectory generation. Validated across diverse Hamiltonian systems, our method in particular improves upon molecular dynamics simulations using machine-learned force fields (MLFF). Our models maintain comparable training and inference cost, but support significantly larger integration timesteps while trained directly on widely-available trajectory-free MLFF datasets.
Assessing generative modeling approaches for free energy estimates in condensed matter
Dec 30, 2025Abstract:The accurate estimation of free energy differences between two states is a long-standing challenge in molecular simulations. Traditional approaches generally rely on sampling multiple intermediate states to ensure sufficient overlap in phase space and are, consequently, computationally expensive. Several generative-model-based methods have recently addressed this challenge by learning a direct bridge between distributions, bypassing the need for intermediate states. However, it remains unclear which approaches provide the best trade-off between efficiency, accuracy, and scalability. In this work, we systematically review these methods and benchmark selected approaches with a focus on condensed-matter systems. In particular, we investigate the performance of discrete and continuous normalizing flows in the context of targeted free energy perturbation as well as FEAT (Free energy Estimators with Adaptive Transport) together with the escorted Jarzynski equality, using coarse-grained monatomic ice and Lennard-Jones solids as benchmark systems. We evaluate accuracy, data efficiency, computational cost, and scalability with system size. Our results provide a quantitative framework for selecting effective free energy estimation strategies in condensed-phase systems.
Operator Forces For Coarse-Grained Molecular Dynamics
Jun 24, 2025Abstract:Coarse-grained (CG) molecular dynamics simulations extend the length and time scale of atomistic simulations by replacing groups of correlated atoms with CG beads. Machine-learned coarse-graining (MLCG) has recently emerged as a promising approach to construct highly accurate force fields for CG molecular dynamics. However, the calibration of MLCG force fields typically hinges on force matching, which demands extensive reference atomistic trajectories with corresponding force labels. In practice, atomistic forces are often not recorded, making traditional force matching infeasible on pre-existing datasets. Recently, noise-based kernels have been introduced to adapt force matching to the low-data regime, including situations in which reference atomistic forces are not present. While this approach produces force fields which recapitulate slow collective motion, it introduces significant local distortions due to the corrupting effects of the noise-based kernel. In this work, we introduce more general kernels based on normalizing flows that substantially reduce these local distortions while preserving global conformational accuracy. We demonstrate our method on small proteins, showing that flow-based kernels can generate high-quality CG forces solely from configurational samples.
Ab-initio simulation of excited-state potential energy surfaces with transferable deep quantum Monte Carlo
Mar 25, 2025Abstract:The accurate quantum chemical calculation of excited states is a challenging task, often requiring computationally demanding methods. When entire ground and excited potential energy surfaces (PESs) are desired, e.g., to predict the interaction of light excitation and structural changes, one is often forced to use cheaper computational methods at the cost of reduced accuracy. Here we introduce a novel method for the geometrically transferable optimization of neural network wave functions that leverages weight sharing and dynamical ordering of electronic states. Our method enables the efficient prediction of ground and excited-state PESs and their intersections at the highest accuracy, demonstrating up to two orders of magnitude cost reduction compared to single-point calculations. We validate our approach on three challenging excited-state PESs, including ethylene, the carbon dimer, and the methylenimmonium cation, indicating that transferable deep-learning QMC can pave the way towards highly accurate simulation of excited-state dynamics.
Doob's Lagrangian: A Sample-Efficient Variational Approach to Transition Path Sampling
Oct 10, 2024



Abstract:Rare event sampling in dynamical systems is a fundamental problem arising in the natural sciences, which poses significant computational challenges due to an exponentially large space of trajectories. For settings where the dynamical system of interest follows a Brownian motion with known drift, the question of conditioning the process to reach a given endpoint or desired rare event is definitively answered by Doob's h-transform. However, the naive estimation of this transform is infeasible, as it requires simulating sufficiently many forward trajectories to estimate rare event probabilities. In this work, we propose a variational formulation of Doob's $h$-transform as an optimization problem over trajectories between a given initial point and the desired ending point. To solve this optimization, we propose a simulation-free training objective with a model parameterization that imposes the desired boundary conditions by design. Our approach significantly reduces the search space over trajectories and avoids expensive trajectory simulation and inefficient importance sampling estimators which are required in existing methods. We demonstrate the ability of our method to find feasible transition paths on real-world molecular simulation and protein folding tasks.
Highly Accurate Real-space Electron Densities with Neural Networks
Sep 02, 2024Abstract:Variational ab-initio methods in quantum chemistry stand out among other methods in providing direct access to the wave function. This allows in principle straightforward extraction of any other observable of interest, besides the energy, but in practice this extraction is often technically difficult and computationally impractical. Here, we consider the electron density as a central observable in quantum chemistry and introduce a novel method to obtain accurate densities from real-space many-electron wave functions by representing the density with a neural network that captures known asymptotic properties and is trained from the wave function by score matching and noise-contrastive estimation. We use variational quantum Monte Carlo with deep-learning ans\"atze (deep QMC) to obtain highly accurate wave functions free of basis set errors, and from them, using our novel method, correspondingly accurate electron densities, which we demonstrate by calculating dipole moments, nuclear forces, contact densities, and other density-based properties.
Artificially intelligent Maxwell's demon for optimal control of open quantum systems
Aug 27, 2024Abstract:Feedback control of open quantum systems is of fundamental importance for practical applications in various contexts, ranging from quantum computation to quantum error correction and quantum metrology. Its use in the context of thermodynamics further enables the study of the interplay between information and energy. However, deriving optimal feedback control strategies is highly challenging, as it involves the optimal control of open quantum systems, the stochastic nature of quantum measurement, and the inclusion of policies that maximize a long-term time- and trajectory-averaged goal. In this work, we employ a reinforcement learning approach to automate and capture the role of a quantum Maxwell's demon: the agent takes the literal role of discovering optimal feedback control strategies in qubit-based systems that maximize a trade-off between measurement-powered cooling and measurement efficiency. Considering weak or projective quantum measurements, we explore different regimes based on the ordering between the thermalization, the measurement, and the unitary feedback timescales, finding different and highly non-intuitive, yet interpretable, strategies. In the thermalization-dominated regime, we find strategies with elaborate finite-time thermalization protocols conditioned on measurement outcomes. In the measurement-dominated regime, we find that optimal strategies involve adaptively measuring different qubit observables reflecting the acquired information, and repeating multiple weak measurements until the quantum state is "sufficiently pure", leading to random walks in state space. Finally, we study the case when all timescales are comparable, finding new feedback control strategies that considerably outperform more intuitive ones. We discuss a two-qubit example where we explore the role of entanglement and conclude discussing the scaling of our results to quantum many-body systems.
Transferable Boltzmann Generators
Jun 20, 2024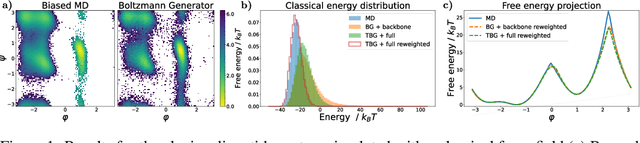
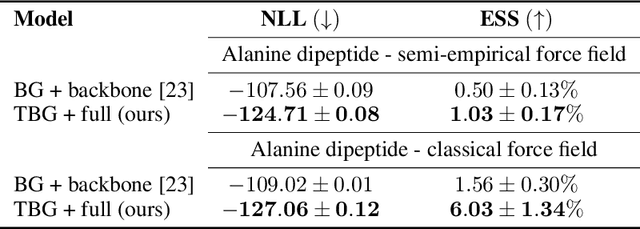
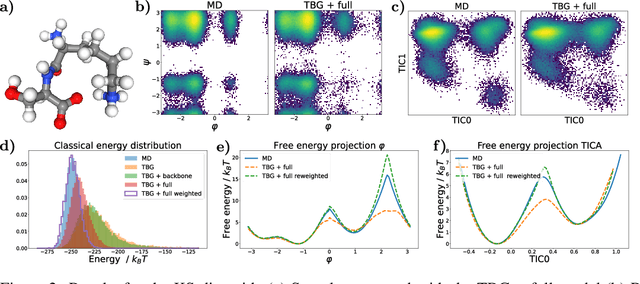
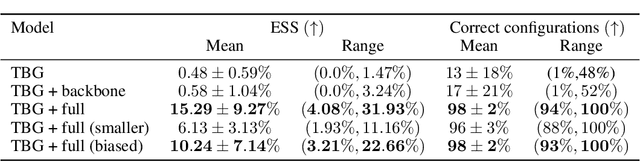
Abstract:The generation of equilibrium samples of molecular systems has been a long-standing problem in statistical physics. Boltzmann Generators are a generative machine learning method that addresses this issue by learning a transformation via a normalizing flow from a simple prior distribution to the target Boltzmann distribution of interest. Recently, flow matching has been employed to train Boltzmann Generators for small molecular systems in Cartesian coordinates. We extend this work and propose a first framework for Boltzmann Generators that are transferable across chemical space, such that they predict zero-shot Boltzmann distributions for test molecules without being retrained for these systems. These transferable Boltzmann Generators allow approximate sampling from the target distribution of unseen systems, as well as efficient reweighting to the target Boltzmann distribution. The transferability of the proposed framework is evaluated on dipeptides, where we show that it generalizes efficiently to unseen systems. Furthermore, we demonstrate that our proposed architecture enhances the efficiency of Boltzmann Generators trained on single molecular systems.
Efficient mapping of phase diagrams with conditional normalizing flows
Jun 18, 2024Abstract:The accurate prediction of phase diagrams is of central importance for both the fundamental understanding of materials as well as for technological applications in material sciences. However, the computational prediction of the relative stability between phases based on their free energy is a daunting task, as traditional free energy estimators require a large amount of simulation data to obtain uncorrelated equilibrium samples over a grid of thermodynamic states. In this work, we develop deep generative machine learning models for entire phase diagrams, employing normalizing flows conditioned on the thermodynamic states, e.g., temperature and pressure, that they map to. By training a single normalizing flow to transform the equilibrium distribution sampled at only one reference thermodynamic state to a wide range of target temperatures and pressures, we can efficiently generate equilibrium samples across the entire phase diagram. Using a permutation-equivariant architecture allows us, thereby, to treat solid and liquid phases on the same footing. We demonstrate our approach by predicting the solid-liquid coexistence line for a Lennard-Jones system in excellent agreement with state-of-the-art free energy methods while significantly reducing the number of energy evaluations needed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge