Shadi Albarqouni
University Hospital Bonn, Venusberg-Campus 1, D-53127, Bonn, Germany, Helmholtz Munich, Ingolstädter Landstraße 1, D-85764, Neuherberg, Germany, Technical University of Munich, Boltzmannstr. 3, D-85748 Garching, Germany
Virtualization of tissue staining in digital pathology using an unsupervised deep learning approach
Oct 15, 2018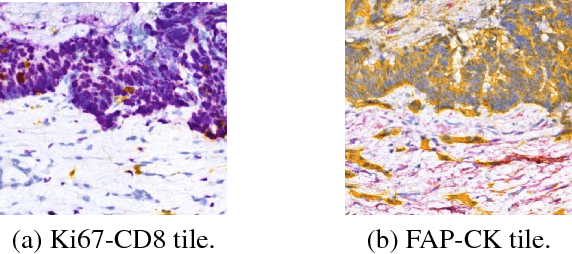
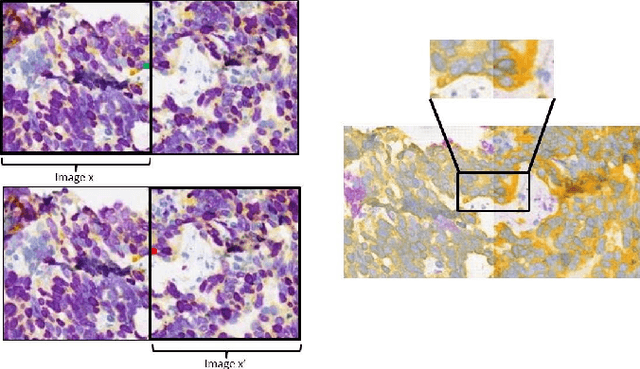

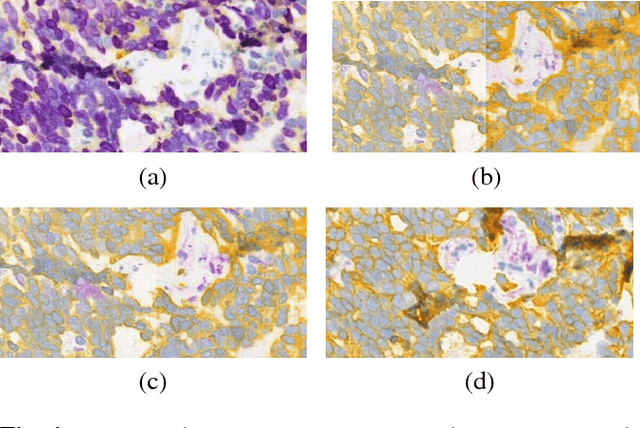
Abstract:Histopathological evaluation of tissue samples is a key practice in patient diagnosis and drug development, especially in oncology. Historically, Hematoxylin and Eosin (H&E) has been used by pathologists as a gold standard staining. However, in many cases, various target specific stains, including immunohistochemistry (IHC), are needed in order to highlight specific structures in the tissue. As tissue is scarce and staining procedures are tedious, it would be beneficial to generate images of stained tissue virtually. Virtual staining could also generate in-silico multiplexing of different stains on the same tissue segment. In this paper, we present a sample application that generates FAP-CK virtual IHC images from Ki67-CD8 real IHC images using an unsupervised deep learning approach based on CycleGAN. We also propose a method to deal with tiling artifacts caused by normalization layers and we validate our approach by comparing the results of tissue analysis algorithms for virtual and real images.
Weakly-Supervised Localization and Classification of Proximal Femur Fractures
Sep 27, 2018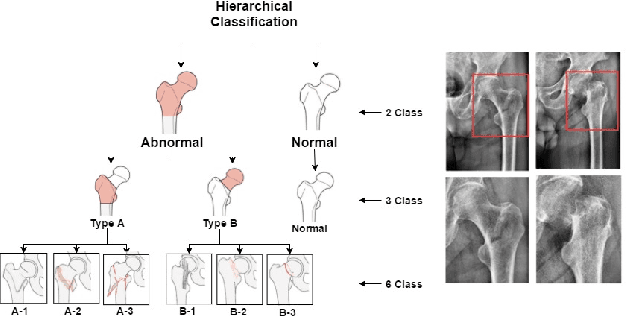
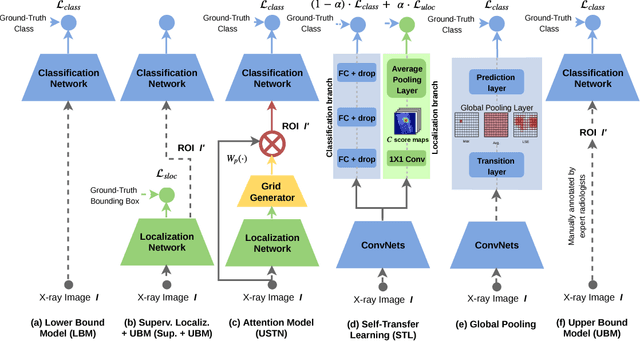
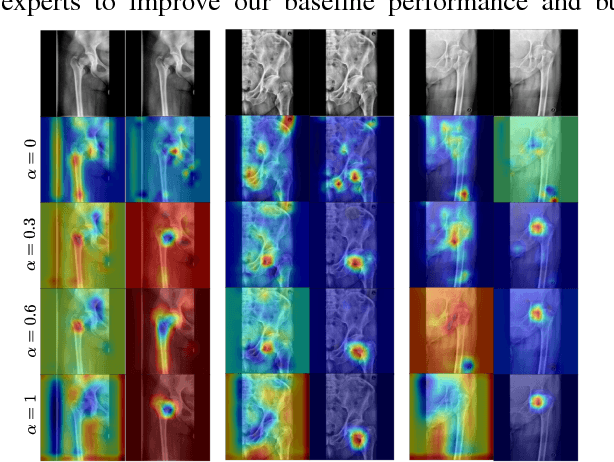
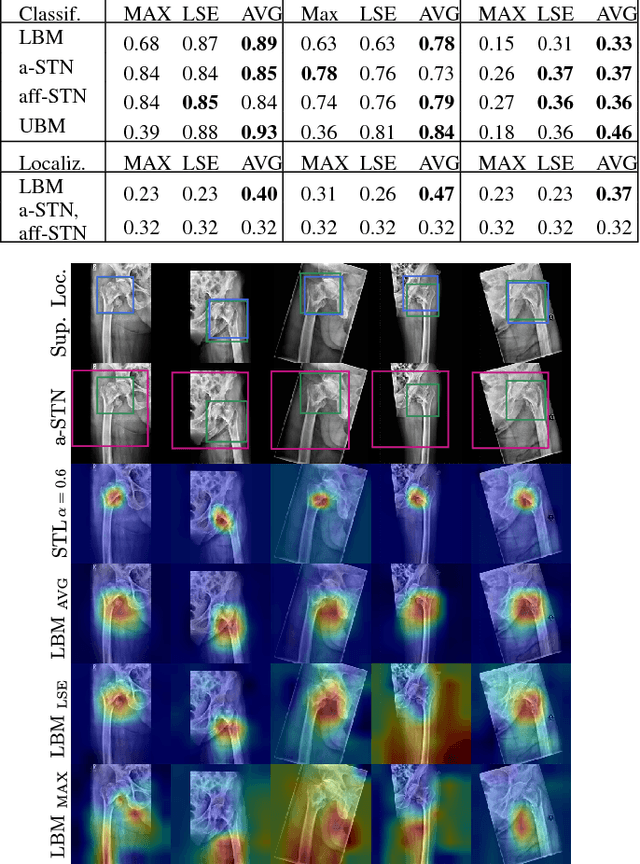
Abstract:In this paper, we target the problem of fracture classification from clinical X-Ray images towards an automated Computer Aided Diagnosis (CAD) system. Although primarily dealing with an image classification problem, we argue that localizing the fracture in the image is crucial to make good class predictions. Therefore, we propose and thoroughly analyze several schemes for simultaneous fracture localization and classification. We show that using an auxiliary localization task, in general, improves the classification performance. Moreover, it is possible to avoid the need for additional localization annotations thanks to recent advancements in weakly-supervised deep learning approaches. Among such approaches, we investigate and adapt Spatial Transformers (ST), Self-Transfer Learning (STL), and localization from global pooling layers. We provide a detailed quantitative and qualitative validation on a dataset of 1347 femur fractures images and report high accuracy with regard to inter-expert correlation values reported in the literature. Our investigations show that i) lesion localization improves the classification outcome, ii) weakly-supervised methods improve baseline classification without any additional cost, iii) STL guides feature activations and boost performance. We plan to make both the dataset and code available.
GANs for Medical Image Analysis
Sep 13, 2018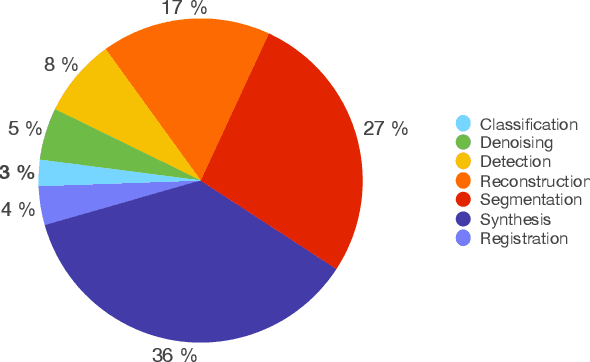

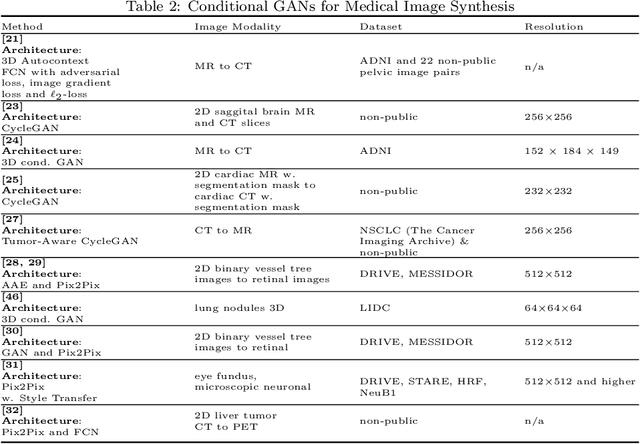
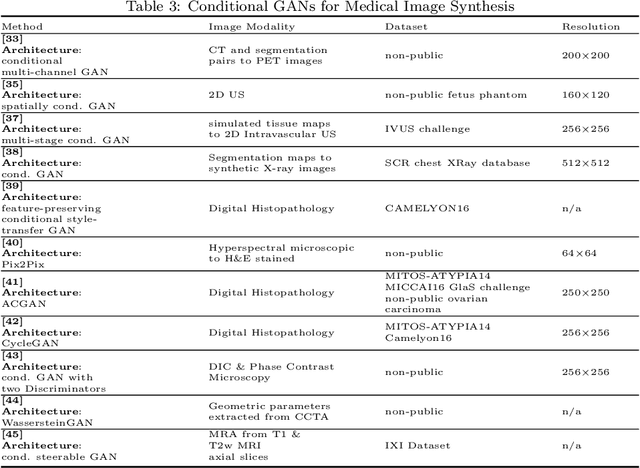
Abstract:Generative Adversarial Networks (GANs) and their extensions have carved open many exciting ways to tackle well known and challenging medical image analysis problems such as medical image denoising, reconstruction, segmentation, data simulation, detection or classification. Furthermore, their ability to synthesize images at unprecedented levels of realism also gives hope that the chronic scarcity of labeled data in the medical field can be resolved with the help of these generative models. In this review paper, a broad overview of recent literature on GANs for medical applications is given, the shortcomings and opportunities of the proposed methods are thoroughly discussed and potential future work is elaborated. A total of 63 papers published until end of July 2018 are reviewed. For quick access, the papers and important details such as the underlying method, datasets and performance are summarized in tables.
Generating Highly Realistic Images of Skin Lesions with GANs
Sep 06, 2018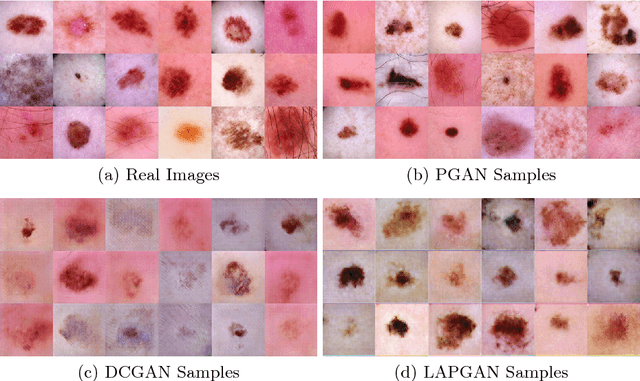
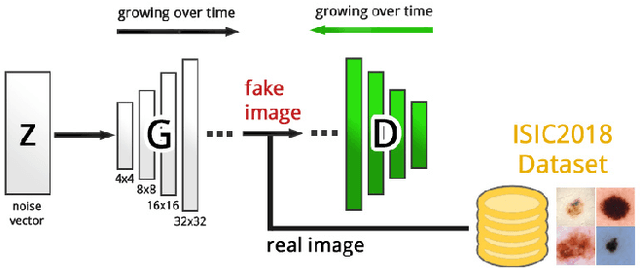
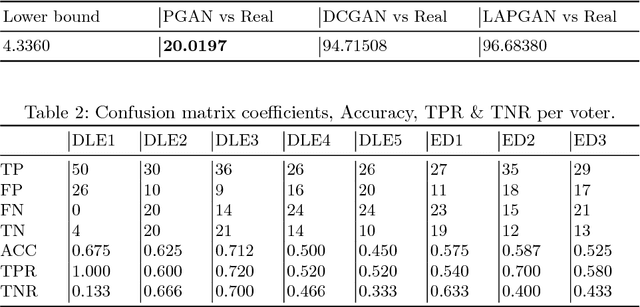
Abstract:As many other machine learning driven medical image analysis tasks, skin image analysis suffers from a chronic lack of labeled data and skewed class distributions, which poses problems for the training of robust and well-generalizing models. The ability to synthesize realistic looking images of skin lesions could act as a reliever for the aforementioned problems. Generative Adversarial Networks (GANs) have been successfully used to synthesize realistically looking medical images, however limited to low resolution, whereas machine learning models for challenging tasks such as skin lesion segmentation or classification benefit from much higher resolution data. In this work, we successfully synthesize realistically looking images of skin lesions with GANs at such high resolution. Therefore, we utilize the concept of progressive growing, which we both quantitatively and qualitatively compare to other GAN architectures such as the DCGAN and the LAPGAN. Our results show that with the help of progressive growing, we can synthesize highly realistic dermoscopic images of skin lesions that even expert dermatologists find hard to distinguish from real ones.
Scene Coordinate and Correspondence Learning for Image-Based Localization
Jul 26, 2018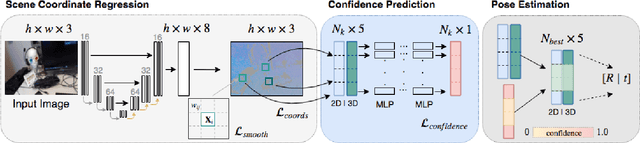
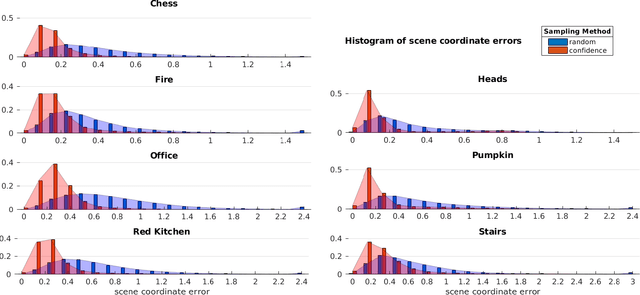
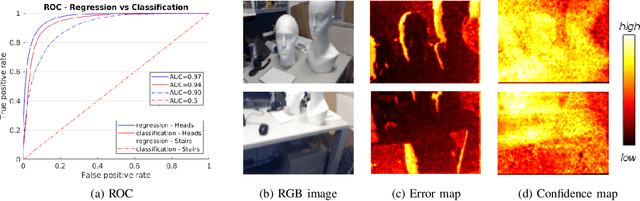
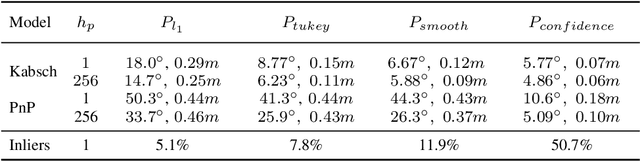
Abstract:Scene coordinate regression has become an essential part of current camera re-localization methods. Different versions, such as regression forests and deep learning methods, have been successfully applied to estimate the corresponding camera pose given a single input image. In this work, we propose to regress the scene coordinates pixel-wise for a given RGB image by using deep learning. Compared to the recent methods, which usually employ RANSAC to obtain a robust pose estimate from the established point correspondences, we propose to regress confidences of these correspondences, which allows us to immediately discard erroneous predictions and improve the initial pose estimates. Finally, the resulting confidences can be used to score initial pose hypothesis and aid in pose refinement, offering a generalized solution to solve this task.
Capsule Networks against Medical Imaging Data Challenges
Jul 19, 2018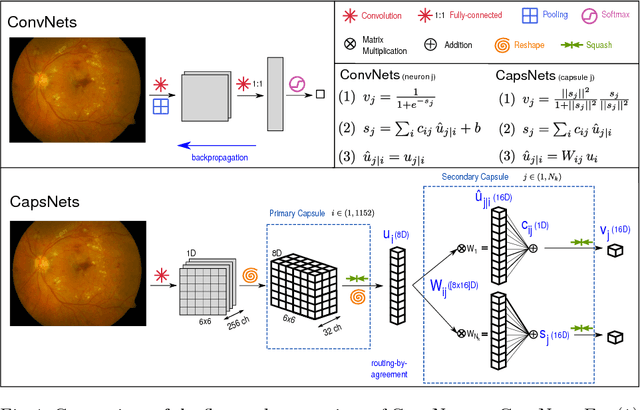
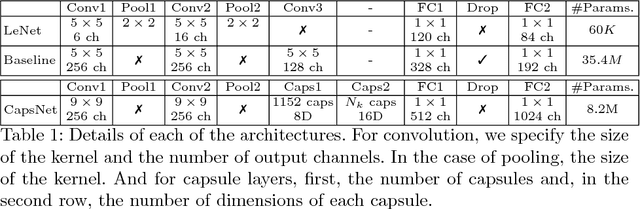
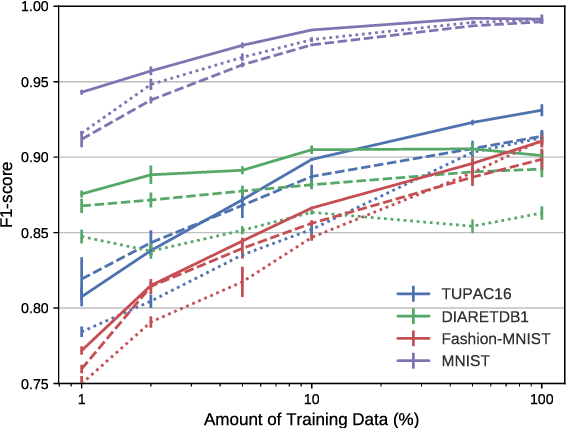
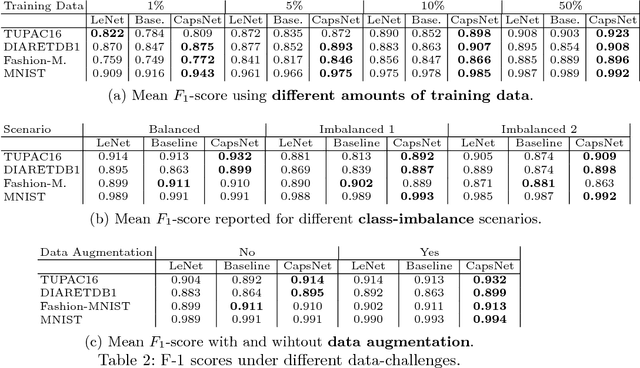
Abstract:A key component to the success of deep learning is the availability of massive amounts of training data. Building and annotating large datasets for solving medical image classification problems is today a bottleneck for many applications. Recently, capsule networks were proposed to deal with shortcomings of Convolutional Neural Networks (ConvNets). In this work, we compare the behavior of capsule networks against ConvNets under typical datasets constraints of medical image analysis, namely, small amounts of annotated data and class-imbalance. We evaluate our experiments on MNIST, Fashion-MNIST and medical (histological and retina images) publicly available datasets. Our results suggest that capsule networks can be trained with less amount of data for the same or better performance and are more robust to an imbalanced class distribution, which makes our approach very promising for the medical imaging community.
When Regression Meets Manifold Learning for Object Recognition and Pose Estimation
May 16, 2018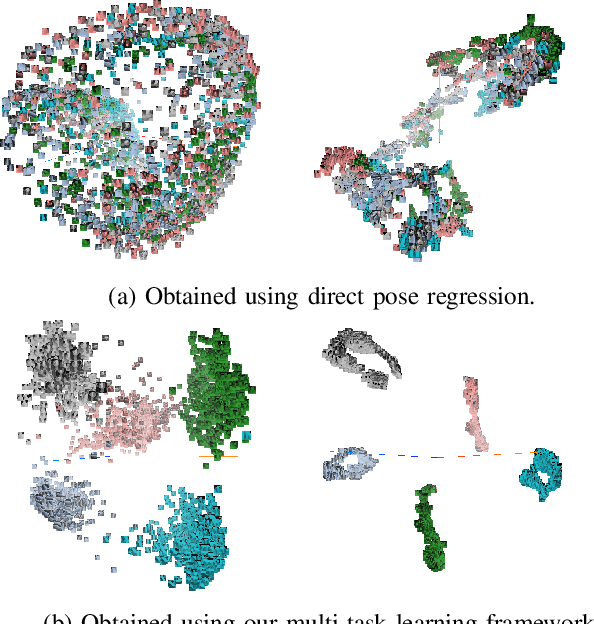
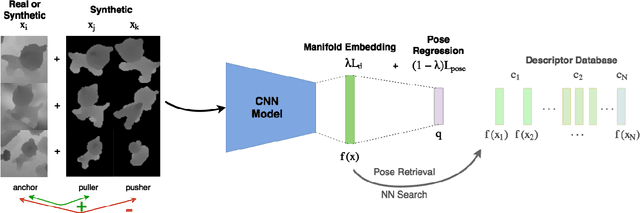
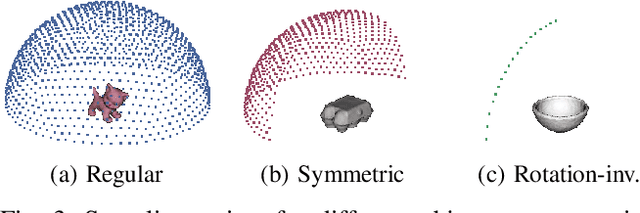
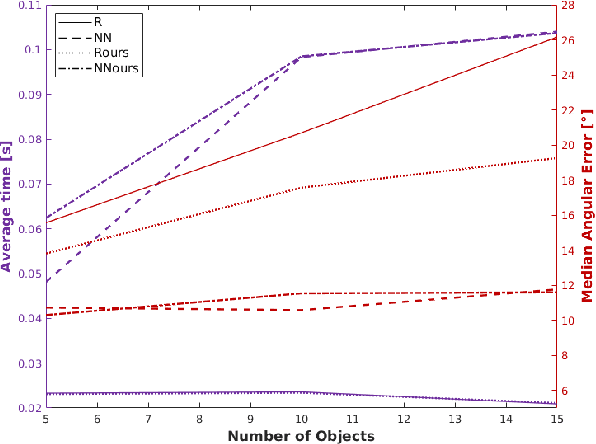
Abstract:In this work, we propose a method for object recognition and pose estimation from depth images using convolutional neural networks. Previous methods addressing this problem rely on manifold learning to learn low dimensional viewpoint descriptors and employ them in a nearest neighbor search on an estimated descriptor space. In comparison we create an efficient multi-task learning framework combining manifold descriptor learning and pose regression. By combining the strengths of manifold learning using triplet loss and pose regression, we could either estimate the pose directly reducing the complexity compared to NN search, or use learned descriptor for the NN descriptor matching. By in depth experimental evaluation of the novel loss function we observed that the view descriptors learned by the network are much more discriminative resulting in almost 30% increase regarding relative pose accuracy compared to related works. On the other hand, regarding directly regressed poses we obtained important improvement compared to simple pose regression. By leveraging the advantages of both manifold learning and regression tasks, we are able to improve the current state-of-the-art for object recognition and pose retrieval that we demonstrate through in depth experimental evaluation.
Multi Layered-Parallel Graph Convolutional Network (ML-PGCN) for Disease Prediction
Apr 28, 2018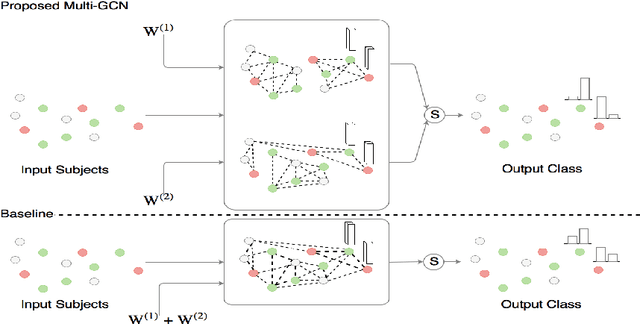
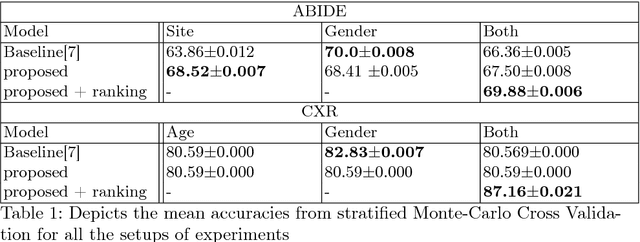
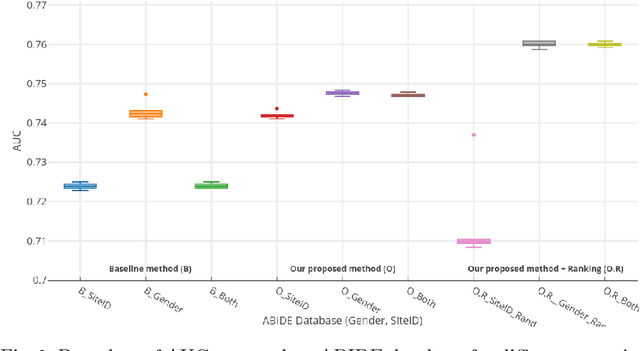
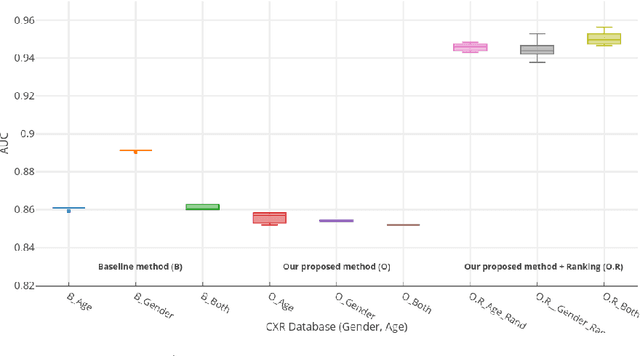
Abstract:Structural data from Electronic Health Records as complementary information to imaging data for disease prediction. We incorporate novel weighting layer into the Graph Convolutional Networks, which weights every element of structural data by exploring its relation to the underlying disease. We demonstrate the superiority of our developed technique in terms of computational speed and obtained encouraging results where our method outperforms the state-of-the-art methods when applied to two publicly available datasets ABIDE and Chest X-ray in terms of relative performance for the accuracy of prediction by 5.31 % and 8.15 % and for the area under the ROC curve by 4.96 % and 10.36 % respectively. Additionally, the model is lightweight, fast and easily trainable.
Domain and Geometry Agnostic CNNs for Left Atrium Segmentation in 3D Ultrasound
Apr 20, 2018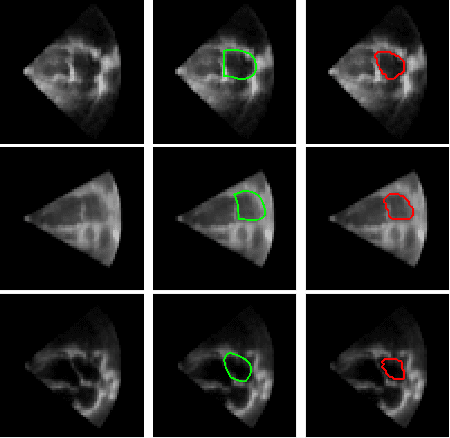

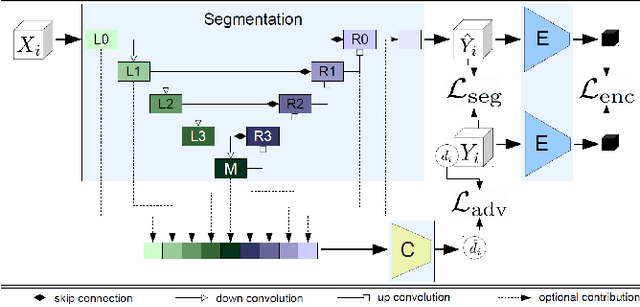
Abstract:Segmentation of the left atrium and deriving its size can help to predict and detect various cardiovascular conditions. Automation of this process in 3D Ultrasound image data is desirable, since manual delineations are time-consuming, challenging and observer-dependent. Convolutional neural networks have made improvements in computer vision and in medical image analysis. They have successfully been applied to segmentation tasks and were extended to work on volumetric data. In this paper we introduce a combined deep-learning based approach on volumetric segmentation in Ultrasound acquisitions with incorporation of prior knowledge about left atrial shape and imaging device. The results show, that including a shape prior helps the domain adaptation and the accuracy of segmentation is further increased with adversarial learning.
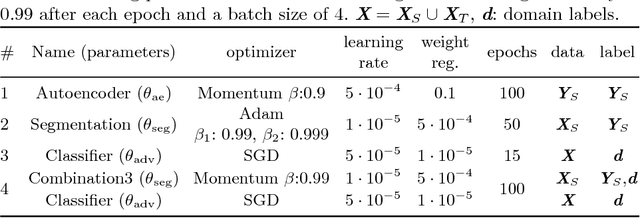
Deep Autoencoding Models for Unsupervised Anomaly Segmentation in Brain MR Images
Apr 12, 2018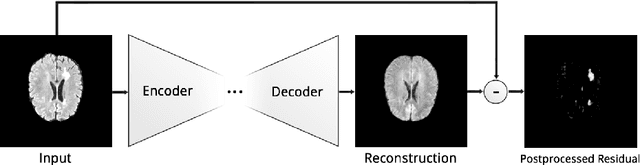
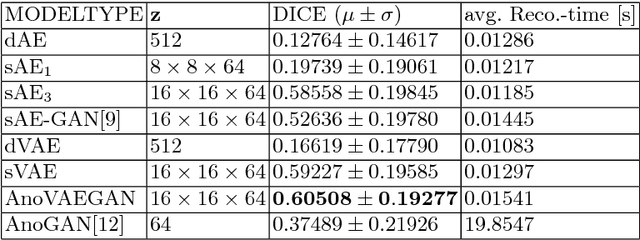
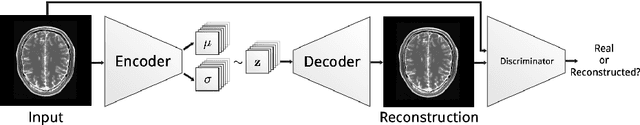
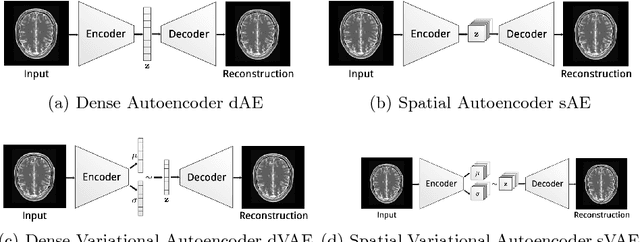
Abstract:Reliably modeling normality and differentiating abnormal appearances from normal cases is a very appealing approach for detecting pathologies in medical images. A plethora of such unsupervised anomaly detection approaches has been made in the medical domain, based on statistical methods, content-based retrieval, clustering and recently also deep learning. Previous approaches towards deep unsupervised anomaly detection model patches of normal anatomy with variants of Autoencoders or GANs, and detect anomalies either as outliers in the learned feature space or from large reconstruction errors. In contrast to these patch-based approaches, we show that deep spatial autoencoding models can be efficiently used to capture normal anatomical variability of entire 2D brain MR images. A variety of experiments on real MR data containing MS lesions corroborates our hypothesis that we can detect and even delineate anomalies in brain MR images by simply comparing input images to their reconstruction. Results show that constraints on the latent space and adversarial training can further improve the segmentation performance over standard deep representation learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge