Satpreet H. Singh
Understanding Electro-communication and Electro-sensing in Weakly Electric Fish using Multi-Agent Deep Reinforcement Learning
Nov 11, 2025Abstract:Weakly electric fish, like Gnathonemus petersii, use a remarkable electrical modality for active sensing and communication, but studying their rich electrosensing and electrocommunication behavior and associated neural activity in naturalistic settings remains experimentally challenging. Here, we present a novel biologically-inspired computational framework to study these behaviors, where recurrent neural network (RNN) based artificial agents trained via multi-agent reinforcement learning (MARL) learn to modulate their electric organ discharges (EODs) and movement patterns to collectively forage in virtual environments. Trained agents demonstrate several emergent features consistent with real fish collectives, including heavy tailed EOD interval distributions, environmental context dependent shifts in EOD interval distributions, and social interaction patterns like freeloading, where agents reduce their EOD rates while benefiting from neighboring agents' active sensing. A minimal two-fish assay further isolates the role of electro-communication, showing that access to conspecific EODs and relative dominance jointly shape foraging success. Notably, these behaviors emerge through evolution-inspired rewards for individual fitness and emergent inter-agent interactions, rather than through rewarding agents explicitly for social interactions. Our work has broad implications for the neuroethology of weakly electric fish, as well as other social, communicating animals in which extensive recordings from multiple individuals, and thus traditional data-driven modeling, are infeasible.
InputDSA: Demixing then Comparing Recurrent and Externally Driven Dynamics
Oct 29, 2025Abstract:In control problems and basic scientific modeling, it is important to compare observations with dynamical simulations. For example, comparing two neural systems can shed light on the nature of emergent computations in the brain and deep neural networks. Recently, Ostrow et al. (2023) introduced Dynamical Similarity Analysis (DSA), a method to measure the similarity of two systems based on their recurrent dynamics rather than geometry or topology. However, DSA does not consider how inputs affect the dynamics, meaning that two similar systems, if driven differently, may be classified as different. Because real-world dynamical systems are rarely autonomous, it is important to account for the effects of input drive. To this end, we introduce a novel metric for comparing both intrinsic (recurrent) and input-driven dynamics, called InputDSA (iDSA). InputDSA extends the DSA framework by estimating and comparing both input and intrinsic dynamic operators using a variant of Dynamic Mode Decomposition with control (DMDc) based on subspace identification. We demonstrate that InputDSA can successfully compare partially observed, input-driven systems from noisy data. We show that when the true inputs are unknown, surrogate inputs can be substituted without a major deterioration in similarity estimates. We apply InputDSA on Recurrent Neural Networks (RNNs) trained with Deep Reinforcement Learning, identifying that high-performing networks are dynamically similar to one another, while low-performing networks are more diverse. Lastly, we apply InputDSA to neural data recorded from rats performing a cognitive task, demonstrating that it identifies a transition from input-driven evidence accumulation to intrinsically-driven decision-making. Our work demonstrates that InputDSA is a robust and efficient method for comparing intrinsic dynamics and the effect of external input on dynamical systems.
Measuring and Controlling Solution Degeneracy across Task-Trained Recurrent Neural Networks
Oct 04, 2024Abstract:Task-trained recurrent neural networks (RNNs) are versatile models of dynamical processes widely used in machine learning and neuroscience. While RNNs are easily trained to perform a wide range of tasks, the nature and extent of the degeneracy in the resultant solutions (i.e., the variability across trained RNNs) remain poorly understood. Here, we provide a unified framework for analyzing degeneracy across three levels: behavior, neural dynamics, and weight space. We analyzed RNNs trained on diverse tasks across machine learning and neuroscience domains, including N-bit flip-flop, sine wave generation, delayed discrimination, and path integration. Our key finding is that the variability across RNN solutions, quantified on the basis of neural dynamics and trained weights, depends primarily on network capacity and task characteristics such as complexity. We introduce information-theoretic measures to quantify task complexity and demonstrate that increasing task complexity consistently reduces degeneracy in neural dynamics and generalization behavior while increasing degeneracy in weight space. These relationships hold across diverse tasks and can be used to control the degeneracy of the solution space of task-trained RNNs. Furthermore, we provide several strategies to control solution degeneracy, enabling task-trained RNNs to learn more consistent or diverse solutions as needed. We envision that these insights will lead to more reliable machine learning models and could inspire strategies to better understand and control degeneracy observed in neuroscience experiments.
A Non-Negative Matrix Factorization Game
Apr 11, 2021



Abstract:We present a novel game-theoretic formulation of Non-Negative Matrix Factorization (NNMF), a popular data-analysis method with many scientific and engineering applications. The game-theoretic formulation is shown to have favorable scaling and parallelization properties, while retaining reconstruction and convergence performance comparable to the traditional Multiplicative Updates algorithm.
Towards naturalistic human neuroscience and neuroengineering: behavior mining in long-term video and neural recordings
Jan 23, 2020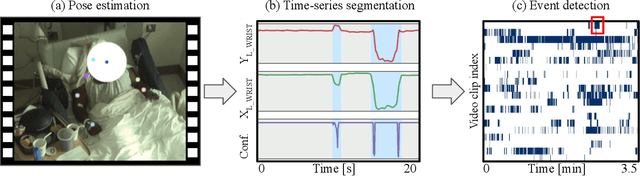
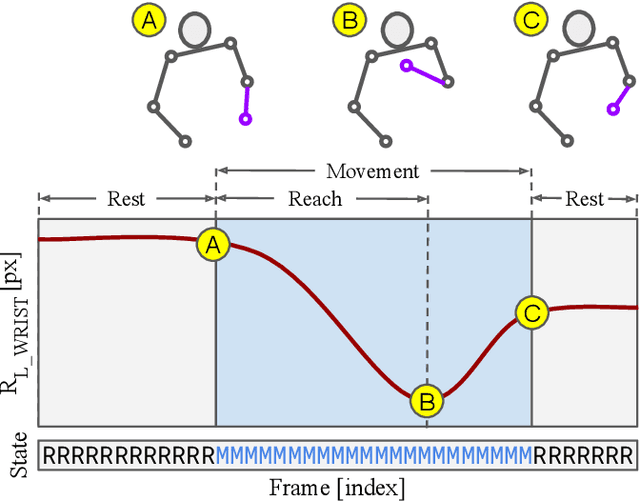
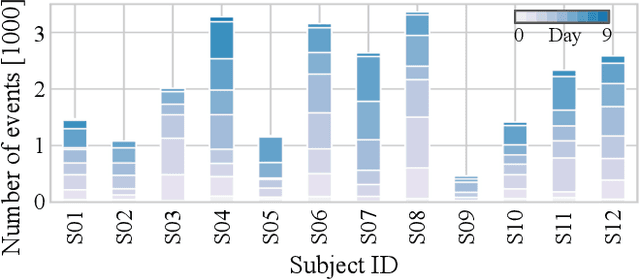

Abstract:Recent advances in brain recording technology and artificial intelligence are propelling a new paradigm in neuroscience beyond the traditional controlled experiment. Naturalistic neuroscience studies neural computations associated with spontaneous behaviors performed in unconstrained settings. Analyzing such unstructured data lacking a priori experimental design remains a significant challenge, especially when the data is multi-modal and long-term. Here we describe an automated approach for analyzing large ($\approx$250 GB/subject) datasets of simultaneously recorded human electrocorticography (ECoG) and naturalistic behavior video data for 12 subjects. Our pipeline discovers and annotates thousands of instances of human upper-limb movement events in long-term (7--9 day) naturalistic behavior data using a combination of computer vision, discrete latent-variable modeling, and string pattern-matching. Analysis of the simultaneously recorded brain data uncovers neural signatures of movement that corroborate prior findings from traditional controlled experiments. We also prototype a decoder for a movement initiation detection task to demonstrate the efficacy of our pipeline as a source of training data for brain-computer interfacing applications. We plan to publish our curated dataset, which captures naturalistic neural and behavioral variability at a scale not previously available. We believe this data will enable further research on models of neural function and decoding that incorporate such naturalistic variability and perform more robustly in real-world settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge