Ronnie Rajan
Cross-Examination Framework: A Task-Agnostic Diagnostic for Information Fidelity in Text-to-Text Generation
Jan 27, 2026Abstract:Traditional metrics like BLEU and BERTScore fail to capture semantic fidelity in generative text-to-text tasks. We adapt the Cross-Examination Framework (CEF) for a reference-free, multi-dimensional evaluation by treating the source and candidate as independent knowledge bases. CEF generates verifiable questions from each text and performs a cross-examination to derive three interpretable scores: Coverage, Conformity, and Consistency. Validated across translation, summarization and clinical note-generation, our framework identifies critical errors, such as content omissions and factual contradictions, missed by standard metrics. A key contribution is a systematic robustness analysis to select a stable judge model. Crucially, the strong correlation between our reference-free and with-reference modes validates CEF's reliability without gold references. Furthermore, human expert validation demonstrates that CEF mismatching questions align with meaning-altering semantic errors higher than with non-semantic errors, particularly excelling at identifying entity-based and relational distortions.
Overalignment in Frontier LLMs: An Empirical Study of Sycophantic Behaviour in Healthcare
Jan 26, 2026Abstract:As LLMs are increasingly integrated into clinical workflows, their tendency for sycophancy, prioritizing user agreement over factual accuracy, poses significant risks to patient safety. While existing evaluations often rely on subjective datasets, we introduce a robust framework grounded in medical MCQA with verifiable ground truths. We propose the Adjusted Sycophancy Score, a novel metric that isolates alignment bias by accounting for stochastic model instability, or "confusability". Through an extensive scaling analysis of the Qwen-3 and Llama-3 families, we identify a clear scaling trajectory for resilience. Furthermore, we reveal a counter-intuitive vulnerability in reasoning-optimized "Thinking" models: while they demonstrate high vanilla accuracy, their internal reasoning traces frequently rationalize incorrect user suggestions under authoritative pressure. Our results across frontier models suggest that benchmark performance is not a proxy for clinical reliability, and that simplified reasoning structures may offer superior robustness against expert-driven sycophancy.
Do Instruction-Tuned Models Always Perform Better Than Base Models? Evidence from Math and Domain-Shifted Benchmarks
Jan 19, 2026Abstract:Instruction finetuning is standard practice for improving LLM performance, yet it remains unclear whether it enhances reasoning or merely induces surface-level pattern matching. We investigate this by evaluating base and instruction-tuned models on standard math benchmarks, structurally perturbed variants, and domain-shifted tasks. Our analysis highlights two key (often overlooked) limitations of instruction tuning. First, the performance advantage is unstable and depends heavily on evaluation settings. In zero-shot CoT settings on GSM8K, base models consistently outperform instruction-tuned variants, with drops as high as 32.67\% (Llama3-70B). Instruction-tuned models only match or exceed this performance when provided with few-shot exemplars, suggesting a reliance on specific prompting patterns rather than intrinsic reasoning. Second, tuning gains are brittle under distribution shift. Our results show that base models surpass instruction-tuned variants on the domain-specific MedCalc benchmark. Additionally, instruction-tuned models show sharp declines on perturbed datasets, indicating sensitivity to prompt structure over robust reasoning.
Named Clinical Entity Recognition Benchmark
Oct 07, 2024



Abstract:This technical report introduces a Named Clinical Entity Recognition Benchmark for evaluating language models in healthcare, addressing the crucial natural language processing (NLP) task of extracting structured information from clinical narratives to support applications like automated coding, clinical trial cohort identification, and clinical decision support. The leaderboard provides a standardized platform for assessing diverse language models, including encoder and decoder architectures, on their ability to identify and classify clinical entities across multiple medical domains. A curated collection of openly available clinical datasets is utilized, encompassing entities such as diseases, symptoms, medications, procedures, and laboratory measurements. Importantly, these entities are standardized according to the Observational Medical Outcomes Partnership (OMOP) Common Data Model, ensuring consistency and interoperability across different healthcare systems and datasets, and a comprehensive evaluation of model performance. Performance of models is primarily assessed using the F1-score, and it is complemented by various assessment modes to provide comprehensive insights into model performance. The report also includes a brief analysis of models evaluated to date, highlighting observed trends and limitations. By establishing this benchmarking framework, the leaderboard aims to promote transparency, facilitate comparative analyses, and drive innovation in clinical entity recognition tasks, addressing the need for robust evaluation methods in healthcare NLP.
MEDIC: Towards a Comprehensive Framework for Evaluating LLMs in Clinical Applications
Sep 11, 2024



Abstract:The rapid development of Large Language Models (LLMs) for healthcare applications has spurred calls for holistic evaluation beyond frequently-cited benchmarks like USMLE, to better reflect real-world performance. While real-world assessments are valuable indicators of utility, they often lag behind the pace of LLM evolution, likely rendering findings obsolete upon deployment. This temporal disconnect necessitates a comprehensive upfront evaluation that can guide model selection for specific clinical applications. We introduce MEDIC, a framework assessing LLMs across five critical dimensions of clinical competence: medical reasoning, ethics and bias, data and language understanding, in-context learning, and clinical safety. MEDIC features a novel cross-examination framework quantifying LLM performance across areas like coverage and hallucination detection, without requiring reference outputs. We apply MEDIC to evaluate LLMs on medical question-answering, safety, summarization, note generation, and other tasks. Our results show performance disparities across model sizes, baseline vs medically finetuned models, and have implications on model selection for applications requiring specific model strengths, such as low hallucination or lower cost of inference. MEDIC's multifaceted evaluation reveals these performance trade-offs, bridging the gap between theoretical capabilities and practical implementation in healthcare settings, ensuring that the most promising models are identified and adapted for diverse healthcare applications.
Med42 -- Evaluating Fine-Tuning Strategies for Medical LLMs: Full-Parameter vs. Parameter-Efficient Approaches
Apr 23, 2024



Abstract:This study presents a comprehensive analysis and comparison of two predominant fine-tuning methodologies - full-parameter fine-tuning and parameter-efficient tuning - within the context of medical Large Language Models (LLMs). We developed and refined a series of LLMs, based on the Llama-2 architecture, specifically designed to enhance medical knowledge retrieval, reasoning, and question-answering capabilities. Our experiments systematically evaluate the effectiveness of these tuning strategies across various well-known medical benchmarks. Notably, our medical LLM Med42 showed an accuracy level of 72% on the US Medical Licensing Examination (USMLE) datasets, setting a new standard in performance for openly available medical LLMs. Through this comparative analysis, we aim to identify the most effective and efficient method for fine-tuning LLMs in the medical domain, thereby contributing significantly to the advancement of AI-driven healthcare applications.
CHAOS Challenge -- Combined (CT-MR) Healthy Abdominal Organ Segmentation
Jan 17, 2020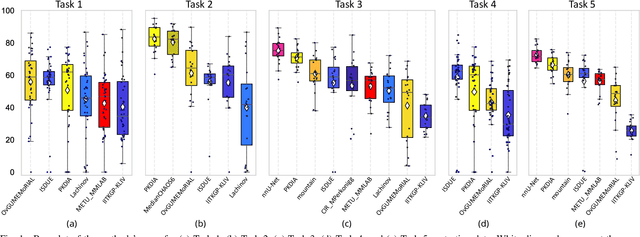
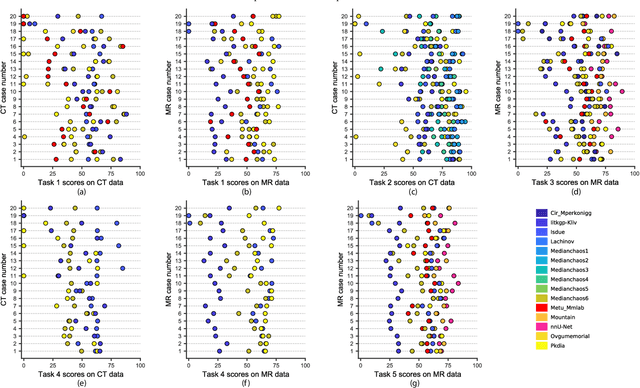
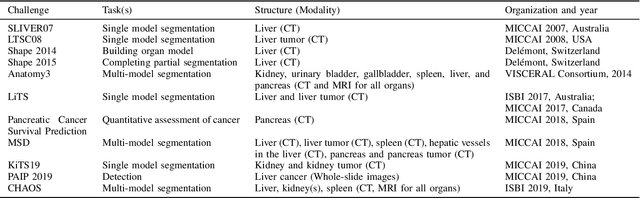
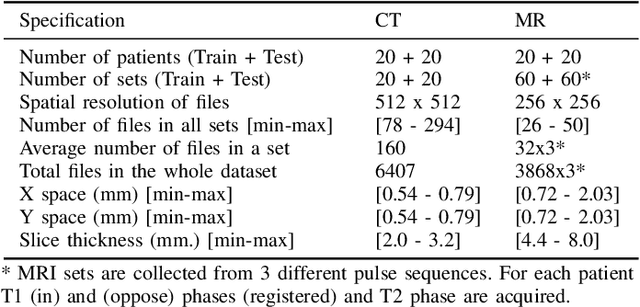
Abstract:Segmentation of abdominal organs has been a comprehensive, yet unresolved, research field for many years. In the last decade, intensive developments in deep learning (DL) have introduced new state-of-the-art segmentation systems. Despite outperforming the overall accuracy of existing systems, the effects of DL model properties and parameters on the performance is hard to interpret. This makes comparative analysis a necessary tool to achieve explainable studies and systems. Moreover, the performance of DL for emerging learning approaches such as cross-modality and multi-modal tasks have been rarely discussed. In order to expand the knowledge in these topics, CHAOS -- Combined (CT-MR) Healthy Abdominal Organ Segmentation challenge has been organized in the IEEE International Symposium on Biomedical Imaging (ISBI), 2019, in Venice, Italy. Despite a large number of the previous abdomen related challenges, the majority of which are focused on tumor/lesion detection and/or classification with a single modality, CHAOS provides both abdominal CT and MR data from healthy subjects. Five different and complementary tasks have been designed to analyze the capabilities of the current approaches from multiple perspectives. The results are investigated thoroughly, compared with manual annotations and interactive methods. The outcomes are reported in detail to reflect the latest advancements in the field. CHAOS challenge and data will be available online to provide a continuous benchmark resource for segmentation.
Adversarially Trained Convolutional Neural Networks for Semantic Segmentation of Ischaemic Stroke Lesion using Multisequence Magnetic Resonance Imaging
Aug 03, 2019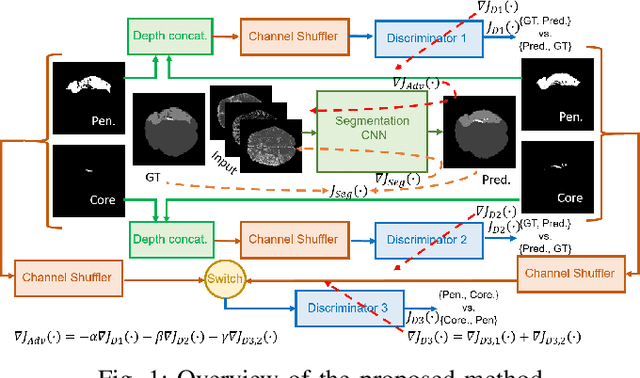
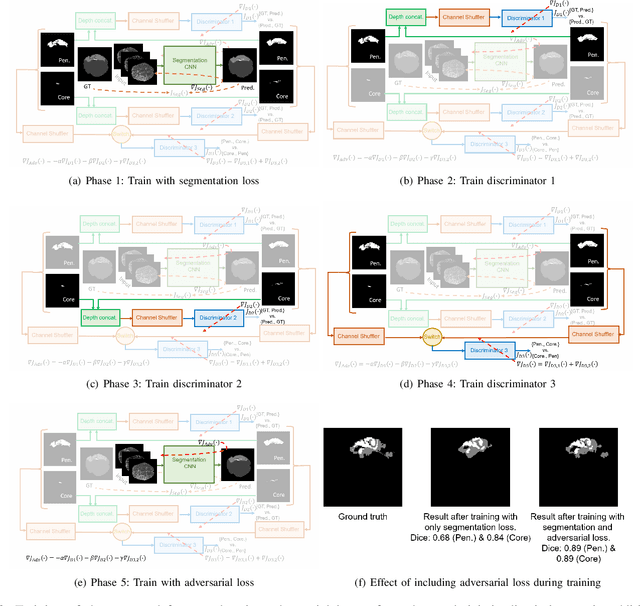
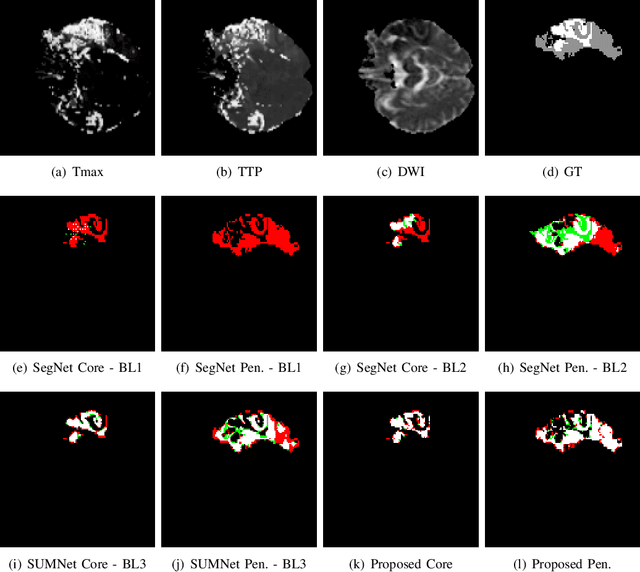
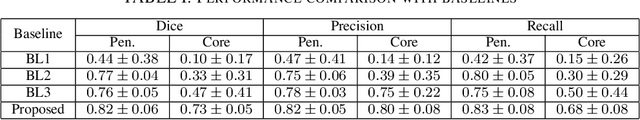
Abstract:Ischaemic stroke is a medical condition caused by occlusion of blood supply to the brain tissue thus forming a lesion. A lesion is zoned into a core associated with irreversible necrosis typically located at the center of the lesion, while reversible hypoxic changes in the outer regions of the lesion are termed as the penumbra. Early estimation of core and penumbra in ischaemic stroke is crucial for timely intervention with thrombolytic therapy to reverse the damage and restore normalcy. Multisequence magnetic resonance imaging (MRI) is commonly employed for clinical diagnosis. However, a sequence singly has not been found to be sufficiently able to differentiate between core and penumbra, while a combination of sequences is required to determine the extent of the damage. The challenge, however, is that with an increase in the number of sequences, it cognitively taxes the clinician to discover symptomatic biomarkers in these images. In this paper, we present a data-driven fully automated method for estimation of core and penumbra in ischaemic lesions using diffusion-weighted imaging (DWI) and perfusion-weighted imaging (PWI) sequence maps of MRI. The method employs recent developments in convolutional neural networks (CNN) for semantic segmentation in medical images. In the absence of availability of a large amount of labeled data, the CNN is trained using an adversarial approach employing cross-entropy as a segmentation loss along with losses aggregated from three discriminators of which two employ relativistic visual Turing test. This method is experimentally validated on the ISLES-2015 dataset through three-fold cross-validation to obtain with an average Dice score of 0.82 and 0.73 for segmentation of penumbra and core respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge