Robin Bülow
MAGO-SP: Detection and Correction of Water-Fat Swaps in Magnitude-Only VIBE MRI
Feb 20, 2025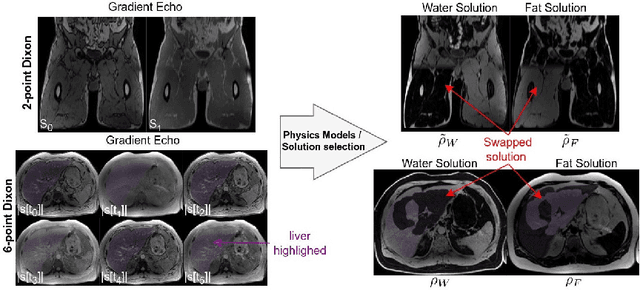
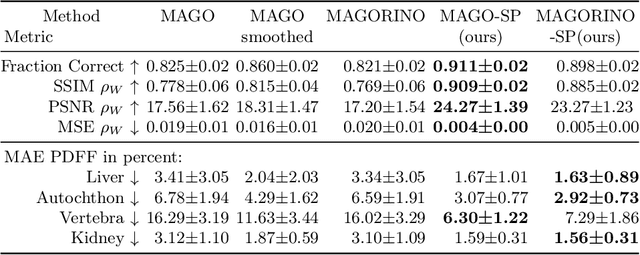
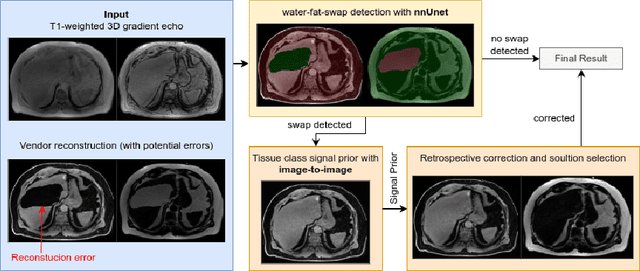
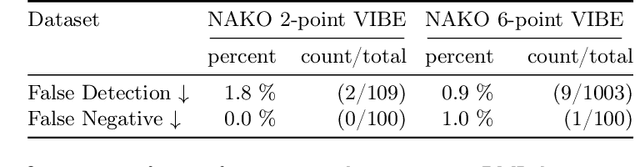
Abstract:Volume Interpolated Breath-Hold Examination (VIBE) MRI generates images suitable for water and fat signal composition estimation. While the two-point VIBE provides water-fat-separated images, the six-point VIBE allows estimation of the effective transversal relaxation rate R2* and the proton density fat fraction (PDFF), which are imaging markers for health and disease. Ambiguity during signal reconstruction can lead to water-fat swaps. This shortcoming challenges the application of VIBE-MRI for automated PDFF analyses of large-scale clinical data and of population studies. This study develops an automated pipeline to detect and correct water-fat swaps in non-contrast-enhanced VIBE images. Our three-step pipeline begins with training a segmentation network to classify volumes as "fat-like" or "water-like," using synthetic water-fat swaps generated by merging fat and water volumes with Perlin noise. Next, a denoising diffusion image-to-image network predicts water volumes as signal priors for correction. Finally, we integrate this prior into a physics-constrained model to recover accurate water and fat signals. Our approach achieves a < 1% error rate in water-fat swap detection for a 6-point VIBE. Notably, swaps disproportionately affect individuals in the Underweight and Class 3 Obesity BMI categories. Our correction algorithm ensures accurate solution selection in chemical phase MRIs, enabling reliable PDFF estimation. This forms a solid technical foundation for automated large-scale population imaging analysis.
TotalVibeSegmentator: Full Torso Segmentation for the NAKO and UK Biobank in Volumetric Interpolated Breath-hold Examination Body Images
May 31, 2024



Abstract:Objectives: To present a publicly available torso segmentation network for large epidemiology datasets on volumetric interpolated breath-hold examination (VIBE) images. Materials & Methods: We extracted preliminary segmentations from TotalSegmentator, spine, and body composition networks for VIBE images, then improved them iteratively and retrained a nnUNet network. Using subsets of NAKO (85 subjects) and UK Biobank (16 subjects), we evaluated with Dice-score on a holdout set (12 subjects) and existing organ segmentation approach (1000 subjects), generating 71 semantic segmentation types for VIBE images. We provide an additional network for the vertebra segments 22 individual vertebra types. Results: We achieved an average Dice score of 0.89 +- 0.07 overall 71 segmentation labels. We scored > 0.90 Dice-score on the abdominal organs except for the pancreas with a Dice of 0.70. Conclusion: Our work offers a detailed and refined publicly available full torso segmentation on VIBE images.
DenseNet and Support Vector Machine classifications of major depressive disorder using vertex-wise cortical features
Nov 18, 2023



Abstract:Major depressive disorder (MDD) is a complex psychiatric disorder that affects the lives of hundreds of millions of individuals around the globe. Even today, researchers debate if morphological alterations in the brain are linked to MDD, likely due to the heterogeneity of this disorder. The application of deep learning tools to neuroimaging data, capable of capturing complex non-linear patterns, has the potential to provide diagnostic and predictive biomarkers for MDD. However, previous attempts to demarcate MDD patients and healthy controls (HC) based on segmented cortical features via linear machine learning approaches have reported low accuracies. In this study, we used globally representative data from the ENIGMA-MDD working group containing an extensive sample of people with MDD (N=2,772) and HC (N=4,240), which allows a comprehensive analysis with generalizable results. Based on the hypothesis that integration of vertex-wise cortical features can improve classification performance, we evaluated the classification of a DenseNet and a Support Vector Machine (SVM), with the expectation that the former would outperform the latter. As we analyzed a multi-site sample, we additionally applied the ComBat harmonization tool to remove potential nuisance effects of site. We found that both classifiers exhibited close to chance performance (balanced accuracy DenseNet: 51%; SVM: 53%), when estimated on unseen sites. Slightly higher classification performance (balanced accuracy DenseNet: 58%; SVM: 55%) was found when the cross-validation folds contained subjects from all sites, indicating site effect. In conclusion, the integration of vertex-wise morphometric features and the use of the non-linear classifier did not lead to the differentiability between MDD and HC. Our results support the notion that MDD classification on this combination of features and classifiers is unfeasible.
An Uncertainty-Aware, Shareable and Transparent Neural Network Architecture for Brain-Age Modeling
Jul 16, 2021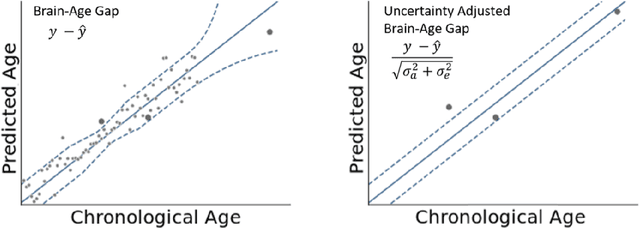
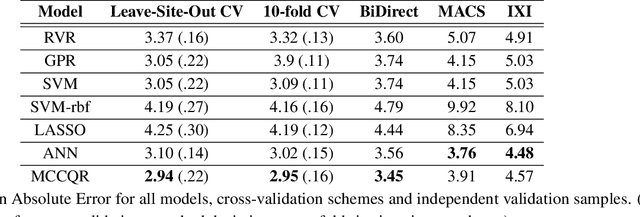
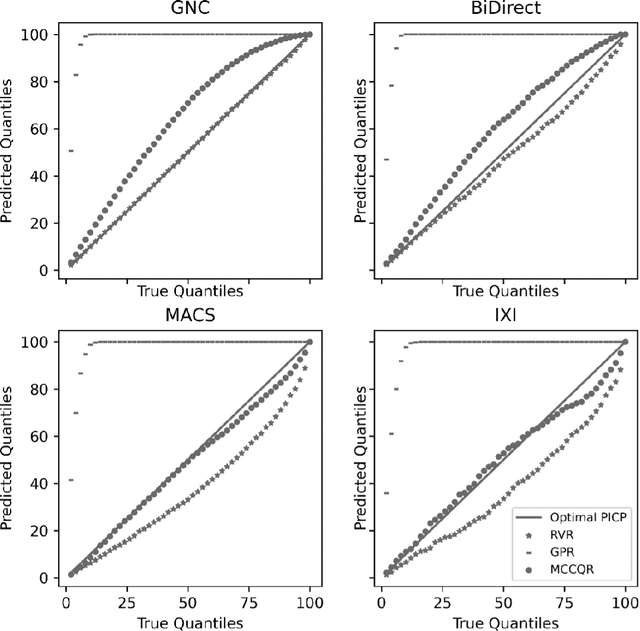
Abstract:The deviation between chronological age and age predicted from neuroimaging data has been identified as a sensitive risk-marker of cross-disorder brain changes, growing into a cornerstone of biological age-research. However, Machine Learning models underlying the field do not consider uncertainty, thereby confounding results with training data density and variability. Also, existing models are commonly based on homogeneous training sets, often not independently validated, and cannot be shared due to data protection issues. Here, we introduce an uncertainty-aware, shareable, and transparent Monte-Carlo Dropout Composite-Quantile-Regression (MCCQR) Neural Network trained on N=10,691 datasets from the German National Cohort. The MCCQR model provides robust, distribution-free uncertainty quantification in high-dimensional neuroimaging data, achieving lower error rates compared to existing models across ten recruitment centers and in three independent validation samples (N=4,004). In two examples, we demonstrate that it prevents spurious associations and increases power to detect accelerated brain-aging. We make the pre-trained model publicly available.
Predicting brain-age from raw T 1 -weighted Magnetic Resonance Imaging data using 3D Convolutional Neural Networks
Mar 22, 2021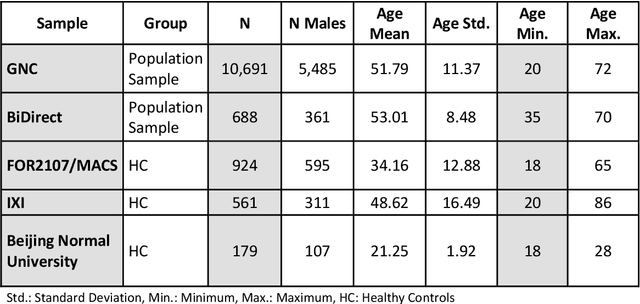
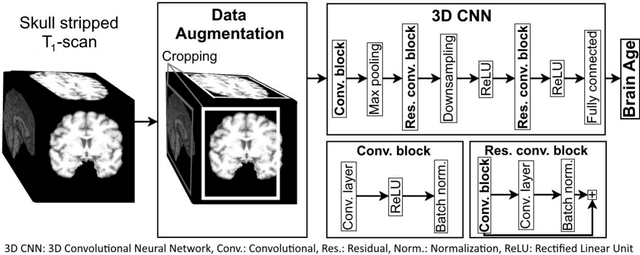
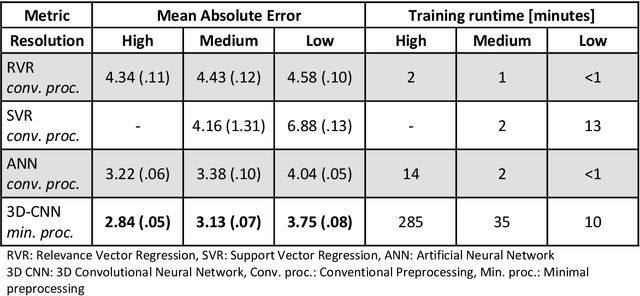
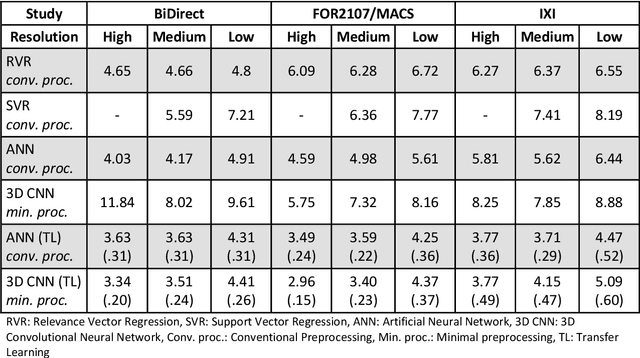
Abstract:Age prediction based on Magnetic Resonance Imaging (MRI) data of the brain is a biomarker to quantify the progress of brain diseases and aging. Current approaches rely on preparing the data with multiple preprocessing steps, such as registering voxels to a standardized brain atlas, which yields a significant computational overhead, hampers widespread usage and results in the predicted brain-age to be sensitive to preprocessing parameters. Here we describe a 3D Convolutional Neural Network (CNN) based on the ResNet architecture being trained on raw, non-registered T$_ 1$-weighted MRI data of N=10,691 samples from the German National Cohort and additionally applied and validated in N=2,173 samples from three independent studies using transfer learning. For comparison, state-of-the-art models using preprocessed neuroimaging data are trained and validated on the same samples. The 3D CNN using raw neuroimaging data predicts age with a mean average deviation of 2.84 years, outperforming the state-of-the-art brain-age models using preprocessed data. Since our approach is invariant to preprocessing software and parameter choices, it enables faster, more robust and more accurate brain-age modeling.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge