Rob J. van der Geest
CMRINet: Joint Groupwise Registration and Segmentation for Cardiac Function Quantification from Cine-MRI
May 22, 2025Abstract:Accurate and efficient quantification of cardiac function is essential for the estimation of prognosis of cardiovascular diseases (CVDs). One of the most commonly used metrics for evaluating cardiac pumping performance is left ventricular ejection fraction (LVEF). However, LVEF can be affected by factors such as inter-observer variability and varying pre-load and after-load conditions, which can reduce its reproducibility. Additionally, cardiac dysfunction may not always manifest as alterations in LVEF, such as in heart failure and cardiotoxicity diseases. An alternative measure that can provide a relatively load-independent quantitative assessment of myocardial contractility is myocardial strain and strain rate. By using LVEF in combination with myocardial strain, it is possible to obtain a thorough description of cardiac function. Automated estimation of LVEF and other volumetric measures from cine-MRI sequences can be achieved through segmentation models, while strain calculation requires the estimation of tissue displacement between sequential frames, which can be accomplished using registration models. These tasks are often performed separately, potentially limiting the assessment of cardiac function. To address this issue, in this study we propose an end-to-end deep learning (DL) model that jointly estimates groupwise (GW) registration and segmentation for cardiac cine-MRI images. The proposed anatomically-guided Deep GW network was trained and validated on a large dataset of 4-chamber view cine-MRI image series of 374 subjects. A quantitative comparison with conventional GW registration using elastix and two DL-based methods showed that the proposed model improved performance and substantially reduced computation time.
Revealing unforeseen diagnostic image features with deep learning by detecting cardiovascular diseases from apical four-chamber ultrasounds
Oct 25, 2021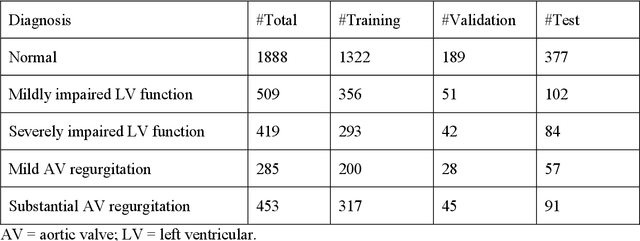
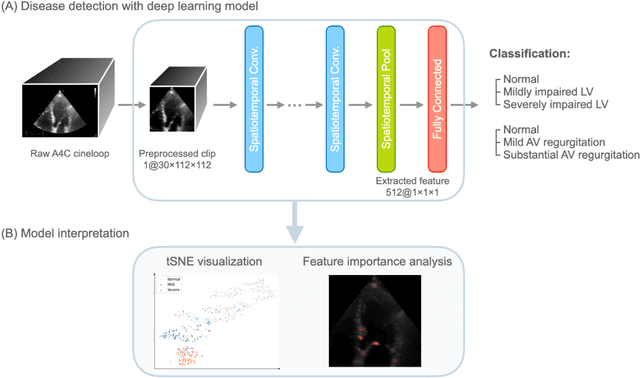
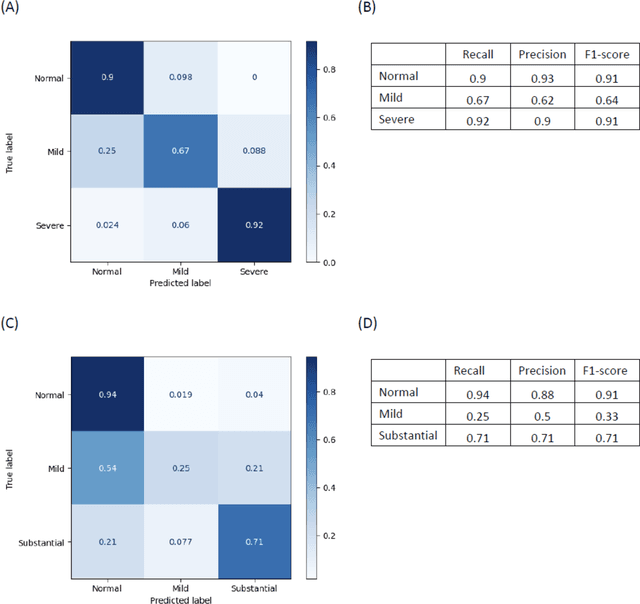
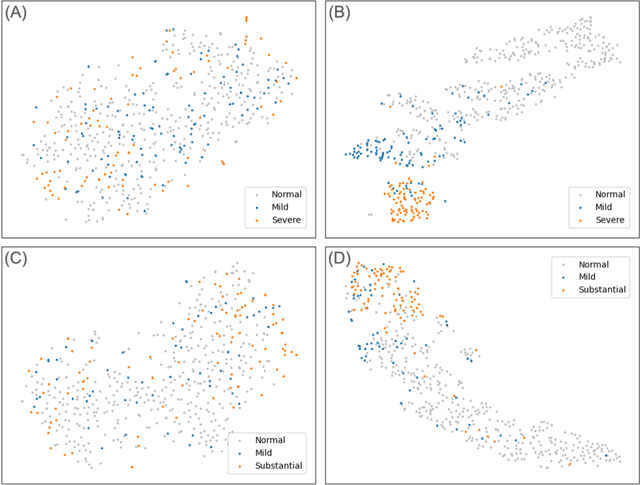
Abstract:Background. With the rise of highly portable, wireless, and low-cost ultrasound devices and automatic ultrasound acquisition techniques, an automated interpretation method requiring only a limited set of views as input could make preliminary cardiovascular disease diagnoses more accessible. In this study, we developed a deep learning (DL) method for automated detection of impaired left ventricular (LV) function and aortic valve (AV) regurgitation from apical four-chamber (A4C) ultrasound cineloops and investigated which anatomical structures or temporal frames provided the most relevant information for the DL model to enable disease classification. Methods and Results. A4C ultrasounds were extracted from 3,554 echocardiograms of patients with either impaired LV function (n=928), AV regurgitation (n=738), or no significant abnormalities (n=1,888). Two convolutional neural networks (CNNs) were trained separately to classify the respective disease cases against normal cases. The overall classification accuracy of the impaired LV function detection model was 86%, and that of the AV regurgitation detection model was 83%. Feature importance analyses demonstrated that the LV myocardium and mitral valve were important for detecting impaired LV function, while the tip of the mitral valve anterior leaflet, during opening, was considered important for detecting AV regurgitation. Conclusion. The proposed method demonstrated the feasibility of a 3D CNN approach in detection of impaired LV function and AV regurgitation using A4C ultrasound cineloops. The current research shows that DL methods can exploit large training data to detect diseases in a different way than conventionally agreed upon methods, and potentially reveal unforeseen diagnostic image features.
Left Ventricle Segmentation via Optical-Flow-Net from Short-axis Cine MRI: Preserving the Temporal Coherence of Cardiac Motion
Oct 20, 2018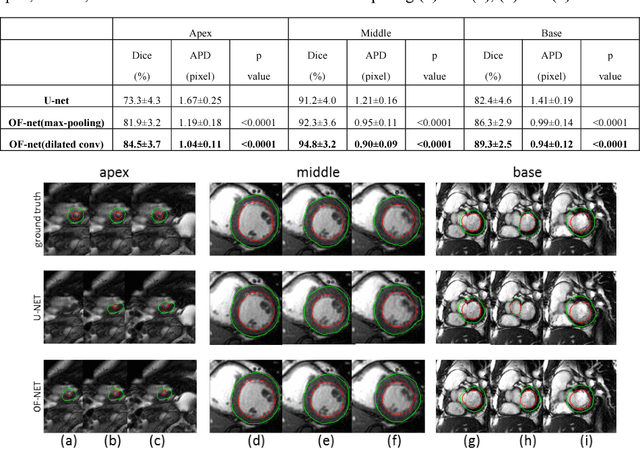
Abstract:Quantitative assessment of left ventricle (LV) function from cine MRI has significant diagnostic and prognostic value for cardiovascular disease patients. The temporal movement of LV provides essential information on the contracting/relaxing pattern of heart, which is keenly evaluated by clinical experts in clinical practice. Inspired by the expert way of viewing Cine MRI, we propose a new CNN module that is able to incorporate the temporal information into LV segmentation from cine MRI. In the proposed CNN, the optical flow (OF) between neighboring frames is integrated and aggregated at feature level, such that temporal coherence in cardiac motion can be taken into account during segmentation. The proposed module is integrated into the U-net architecture without need of additional training. Furthermore, dilated convolution is introduced to improve the spatial accuracy of segmentation. Trained and tested on the Cardiac Atlas database, the proposed network resulted in a Dice index of 95% and an average perpendicular distance of 0.9 pixels for the middle LV contour, significantly outperforming the original U-net that processes each frame individually. Notably, the proposed method improved the temporal coherence of LV segmentation results, especially at the LV apex and base where the cardiac motion is difficult to follow.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge