Quan Wen
From Pixels to Views: Learning Angular-Aware and Physics-Consistent Representations for Light Field Microscopy
Oct 26, 2025Abstract:Light field microscopy (LFM) has become an emerging tool in neuroscience for large-scale neural imaging in vivo, notable for its single-exposure volumetric imaging, broad field of view, and high temporal resolution. However, learning-based 3D reconstruction in XLFM remains underdeveloped due to two core challenges: the absence of standardized datasets and the lack of methods that can efficiently model its angular-spatial structure while remaining physically grounded. We address these challenges by introducing three key contributions. First, we construct the XLFM-Zebrafish benchmark, a large-scale dataset and evaluation suite for XLFM reconstruction. Second, we propose Masked View Modeling for Light Fields (MVN-LF), a self-supervised task that learns angular priors by predicting occluded views, improving data efficiency. Third, we formulate the Optical Rendering Consistency Loss (ORC Loss), a differentiable rendering constraint that enforces alignment between predicted volumes and their PSF-based forward projections. On the XLFM-Zebrafish benchmark, our method improves PSNR by 7.7% over state-of-the-art baselines.
A Survey on Graph Classification and Link Prediction based on GNN
Jul 03, 2023



Abstract:Traditional convolutional neural networks are limited to handling Euclidean space data, overlooking the vast realm of real-life scenarios represented as graph data, including transportation networks, social networks, and reference networks. The pivotal step in transferring convolutional neural networks to graph data analysis and processing lies in the construction of graph convolutional operators and graph pooling operators. This comprehensive review article delves into the world of graph convolutional neural networks. Firstly, it elaborates on the fundamentals of graph convolutional neural networks. Subsequently, it elucidates the graph neural network models based on attention mechanisms and autoencoders, summarizing their application in node classification, graph classification, and link prediction along with the associated datasets.
Rapid detection and recognition of whole brain activity in a freely behaving Caenorhabditis elegans
Sep 23, 2021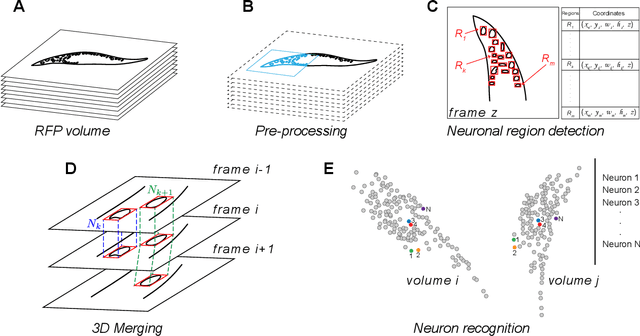

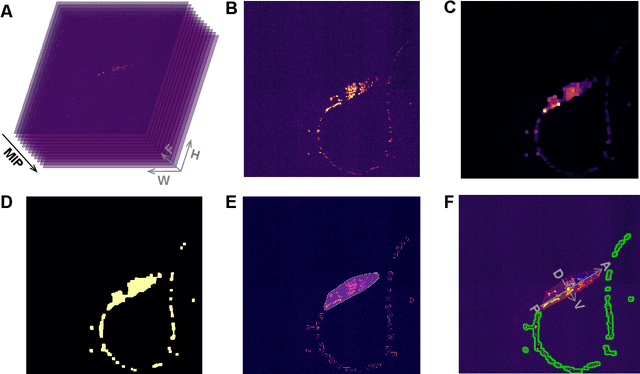
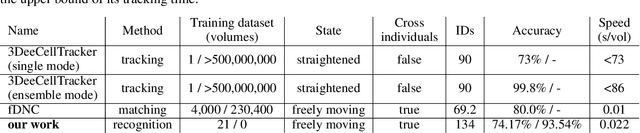
Abstract:Advanced volumetric imaging methods and genetically encoded activity indicators have permitted a comprehensive characterization of whole brain activity at single neuron resolution in \textit{Caenorhabditis elegans}. The constant motion and deformation of the mollusc nervous system, however, impose a great challenge for a consistent identification of densely packed neurons in a behaving animal. Here, we propose a cascade solution for long-term and rapid recognition of head ganglion neurons in a freely moving \textit{C. elegans}. First, potential neuronal regions from a stack of fluorescence images are detected by a deep learning algorithm. Second, 2 dimensional neuronal regions are fused into 3 dimensional neuron entities. Third, by exploiting the neuronal density distribution surrounding a neuron and relative positional information between neurons, a multi-class artificial neural network transforms engineered neuronal feature vectors into digital neuronal identities. Under the constraint of a small number (20-40 volumes) of training samples, our bottom-up approach is able to process each volume - $1024 \times 1024 \times 18$ in voxels - in less than 1 second and achieves an accuracy of $91\%$ in neuronal detection and $74\%$ in neuronal recognition. Our work represents an important development towards a rapid and fully automated algorithm for decoding whole brain activity underlying natural animal behaviors.
Lesion Border Detection in Dermoscopy Images Using Ensembles of Thresholding Methods
Dec 26, 2013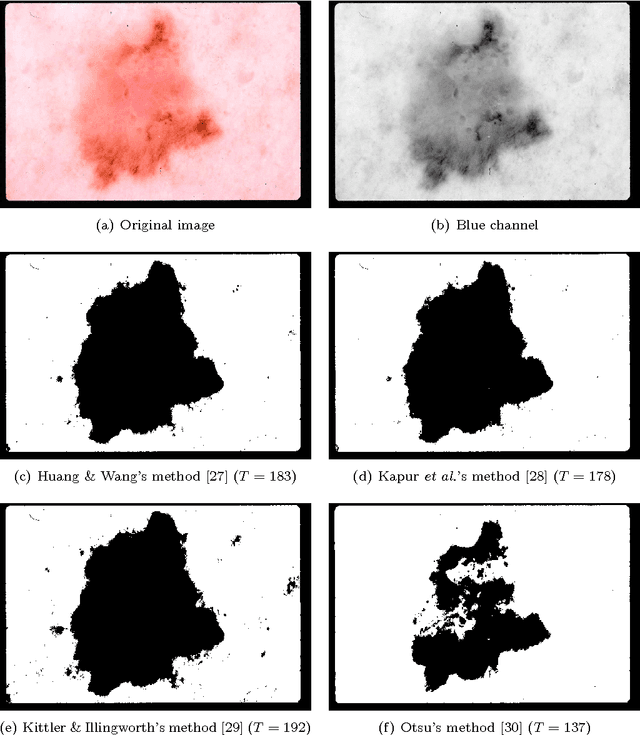
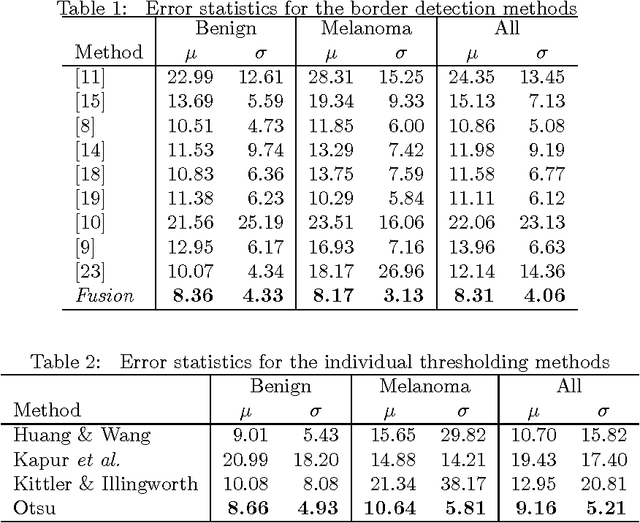
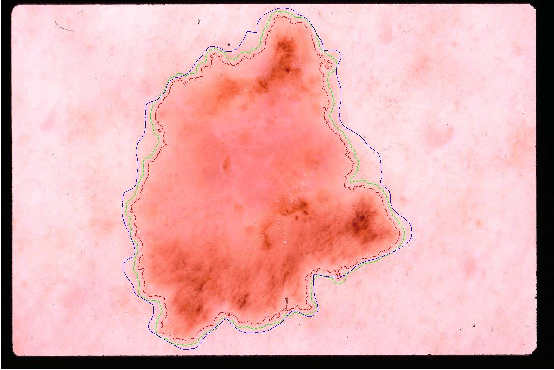
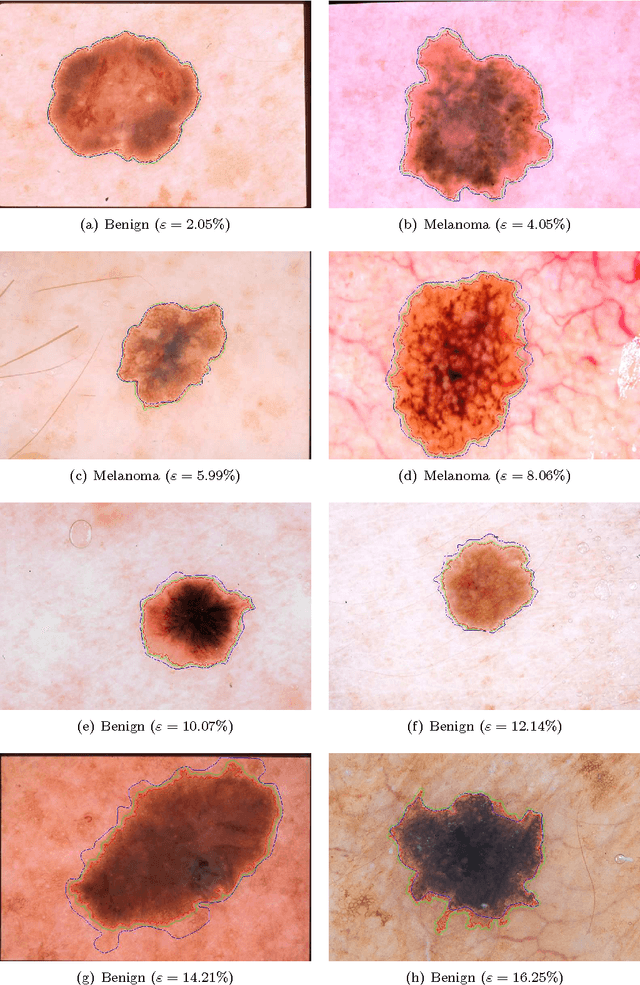
Abstract:Dermoscopy is one of the major imaging modalities used in the diagnosis of melanoma and other pigmented skin lesions. Due to the difficulty and subjectivity of human interpretation, automated analysis of dermoscopy images has become an important research area. Border detection is often the first step in this analysis. In many cases, the lesion can be roughly separated from the background skin using a thresholding method applied to the blue channel. However, no single thresholding method appears to be robust enough to successfully handle the wide variety of dermoscopy images encountered in clinical practice. In this paper, we present an automated method for detecting lesion borders in dermoscopy images using ensembles of thresholding methods. Experiments on a difficult set of 90 images demonstrate that the proposed method is robust, fast, and accurate when compared to nine state-of-the-art methods.
* 8 pages, 3 figures, 2 tables. arXiv admin note: substantial text overlap with arXiv:1009.1362
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge