Prathosh AP
MeasureNet: Measurement Based Celiac Disease Identification
Dec 02, 2024



Abstract:Celiac disease is an autoimmune disorder triggered by the consumption of gluten. It causes damage to the villi, the finger-like projections in the small intestine that are responsible for nutrient absorption. Additionally, the crypts, which form the base of the villi, are also affected, impairing the regenerative process. The deterioration in villi length, computed as the villi-to-crypt length ratio, indicates the severity of celiac disease. However, manual measurement of villi-crypt length can be both time-consuming and susceptible to inter-observer variability, leading to inconsistencies in diagnosis. While some methods can perform measurement as a post-hoc process, they are prone to errors in the initial stages. This gap underscores the need for pathologically driven solutions that enhance measurement accuracy and reduce human error in celiac disease assessments. Our proposed method, MeasureNet, is a pathologically driven polyline detection framework incorporating polyline localization and object-driven losses specifically designed for measurement tasks. Furthermore, we leverage segmentation model to provide auxiliary guidance about crypt location when crypt are partially visible. To ensure that model is not overdependent on segmentation mask we enhance model robustness through a mask feature mixup technique. Additionally, we introduce a novel dataset for grading celiac disease, consisting of 750 annotated duodenum biopsy images. MeasureNet achieves an 82.66% classification accuracy for binary classification and 81% accuracy for multi-class grading of celiac disease. Code: https://github.com/dair-iitd/MeasureNet
ReMOVE: A Reference-free Metric for Object Erasure
Sep 01, 2024



Abstract:We introduce $\texttt{ReMOVE}$, a novel reference-free metric for assessing object erasure efficacy in diffusion-based image editing models post-generation. Unlike existing measures such as LPIPS and CLIPScore, $\texttt{ReMOVE}$ addresses the challenge of evaluating inpainting without a reference image, common in practical scenarios. It effectively distinguishes between object removal and replacement. This is a key issue in diffusion models due to stochastic nature of image generation. Traditional metrics fail to align with the intuitive definition of inpainting, which aims for (1) seamless object removal within masked regions (2) while preserving the background continuity. $\texttt{ReMOVE}$ not only correlates with state-of-the-art metrics and aligns with human perception but also captures the nuanced aspects of the inpainting process, providing a finer-grained evaluation of the generated outputs.
AdaKD: Dynamic Knowledge Distillation of ASR models using Adaptive Loss Weighting
May 11, 2024


Abstract:Knowledge distillation, a widely used model compression technique, works on the basis of transferring knowledge from a cumbersome teacher model to a lightweight student model. The technique involves jointly optimizing the task specific and knowledge distillation losses with a weight assigned to them. Despite these weights playing a crucial role in the performance of the distillation process, current methods provide equal weight to both losses, leading to suboptimal performance. In this paper, we propose Adaptive Knowledge Distillation, a novel technique inspired by curriculum learning to adaptively weigh the losses at instance level. This technique goes by the notion that sample difficulty increases with teacher loss. Our method follows a plug-and-play paradigm that can be applied on top of any task-specific and distillation objectives. Experiments show that our method performs better than conventional knowledge distillation method and existing instance-level loss functions.
WISER: Weak supervISion and supErvised Representation learning to improve drug response prediction in cancer
May 07, 2024Abstract:Cancer, a leading cause of death globally, occurs due to genomic changes and manifests heterogeneously across patients. To advance research on personalized treatment strategies, the effectiveness of various drugs on cells derived from cancers (`cell lines') is experimentally determined in laboratory settings. Nevertheless, variations in the distribution of genomic data and drug responses between cell lines and humans arise due to biological and environmental differences. Moreover, while genomic profiles of many cancer patients are readily available, the scarcity of corresponding drug response data limits the ability to train machine learning models that can predict drug response in patients effectively. Recent cancer drug response prediction methods have largely followed the paradigm of unsupervised domain-invariant representation learning followed by a downstream drug response classification step. Introducing supervision in both stages is challenging due to heterogeneous patient response to drugs and limited drug response data. This paper addresses these challenges through a novel representation learning method in the first phase and weak supervision in the second. Experimental results on real patient data demonstrate the efficacy of our method (WISER) over state-of-the-art alternatives on predicting personalized drug response.
LoMOE: Localized Multi-Object Editing via Multi-Diffusion
Mar 01, 2024



Abstract:Recent developments in the field of diffusion models have demonstrated an exceptional capacity to generate high-quality prompt-conditioned image edits. Nevertheless, previous approaches have primarily relied on textual prompts for image editing, which tend to be less effective when making precise edits to specific objects or fine-grained regions within a scene containing single/multiple objects. We introduce a novel framework for zero-shot localized multi-object editing through a multi-diffusion process to overcome this challenge. This framework empowers users to perform various operations on objects within an image, such as adding, replacing, or editing $\textbf{many}$ objects in a complex scene $\textbf{in one pass}$. Our approach leverages foreground masks and corresponding simple text prompts that exert localized influences on the target regions resulting in high-fidelity image editing. A combination of cross-attention and background preservation losses within the latent space ensures that the characteristics of the object being edited are preserved while simultaneously achieving a high-quality, seamless reconstruction of the background with fewer artifacts compared to the current methods. We also curate and release a dataset dedicated to multi-object editing, named $\texttt{LoMOE}$-Bench. Our experiments against existing state-of-the-art methods demonstrate the improved effectiveness of our approach in terms of both image editing quality and inference speed.
FAST: Feature Aware Similarity Thresholding for Weak Unlearning in Black-Box Generative Models
Dec 22, 2023



Abstract:The heightened emphasis on the regulation of deep generative models, propelled by escalating concerns pertaining to privacy and compliance with regulatory frameworks, underscores the imperative need for precise control mechanisms over these models. This urgency is particularly underscored by instances in which generative models generate outputs that encompass objectionable, offensive, or potentially injurious content. In response, machine unlearning has emerged to selectively forget specific knowledge or remove the influence of undesirable data subsets from pre-trained models. However, modern machine unlearning approaches typically assume access to model parameters and architectural details during unlearning, which is not always feasible. In multitude of downstream tasks, these models function as black-box systems, with inaccessible pre-trained parameters, architectures, and training data. In such scenarios, the possibility of filtering undesired outputs becomes a practical alternative. The primary goal of this study is twofold: first, to elucidate the relationship between filtering and unlearning processes, and second, to formulate a methodology aimed at mitigating the display of undesirable outputs generated from models characterized as black-box systems. Theoretical analysis in this study demonstrates that, in the context of black-box models, filtering can be seen as a form of weak unlearning. Our proposed \textbf{\textit{Feature Aware Similarity Thresholding(FAST)}} method effectively suppresses undesired outputs by systematically encoding the representation of unwanted features in the latent space.
Fusing Conditional Submodular GAN and Programmatic Weak Supervision
Dec 16, 2023Abstract:Programmatic Weak Supervision (PWS) and generative models serve as crucial tools that enable researchers to maximize the utility of existing datasets without resorting to laborious data gathering and manual annotation processes. PWS uses various weak supervision techniques to estimate the underlying class labels of data, while generative models primarily concentrate on sampling from the underlying distribution of the given dataset. Although these methods have the potential to complement each other, they have mostly been studied independently. Recently, WSGAN proposed a mechanism to fuse these two models. Their approach utilizes the discrete latent factors of InfoGAN to train the label model and leverages the class-dependent information of the label model to generate images of specific classes. However, the disentangled latent factors learned by InfoGAN might not necessarily be class-specific and could potentially affect the label model's accuracy. Moreover, prediction made by the label model is often noisy in nature and can have a detrimental impact on the quality of images generated by GAN. In our work, we address these challenges by (i) implementing a noise-aware classifier using the pseudo labels generated by the label model (ii) utilizing the noise-aware classifier's prediction to train the label model and generate class-conditional images. Additionally, we also investigate the effect of training the classifier with a subset of the dataset within a defined uncertainty budget on pseudo labels. We accomplish this by formalizing the subset selection problem as a submodular maximization objective with a knapsack constraint on the entropy of pseudo labels. We conduct experiments on multiple datasets and demonstrate the efficacy of our methods on several tasks vis-a-vis the current state-of-the-art methods.
DeGPR: Deep Guided Posterior Regularization for Multi-Class Cell Detection and Counting
Apr 03, 2023Abstract:Multi-class cell detection and counting is an essential task for many pathological diagnoses. Manual counting is tedious and often leads to inter-observer variations among pathologists. While there exist multiple, general-purpose, deep learning-based object detection and counting methods, they may not readily transfer to detecting and counting cells in medical images, due to the limited data, presence of tiny overlapping objects, multiple cell types, severe class-imbalance, minute differences in size/shape of cells, etc. In response, we propose guided posterior regularization (DeGPR), which assists an object detector by guiding it to exploit discriminative features among cells. The features may be pathologist-provided or inferred directly from visual data. We validate our model on two publicly available datasets (CoNSeP and MoNuSAC), and on MuCeD, a novel dataset that we contribute. MuCeD consists of 55 biopsy images of the human duodenum for predicting celiac disease. We perform extensive experimentation with three object detection baselines on three datasets to show that DeGPR is model-agnostic, and consistently improves baselines obtaining up to 9% (absolute) mAP gains.
Discovering mesoscopic descriptions of collective movement with neural stochastic modelling
Mar 17, 2023



Abstract:Collective motion is an ubiquitous phenomenon in nature, inspiring engineers, physicists and mathematicians to develop mathematical models and bio-inspired designs. Collective motion at small to medium group sizes ($\sim$10-1000 individuals, also called the `mesoscale'), can show nontrivial features due to stochasticity. Therefore, characterizing both the deterministic and stochastic aspects of the dynamics is crucial in the study of mesoscale collective phenomena. Here, we use a physics-inspired, neural-network based approach to characterize the stochastic group dynamics of interacting individuals, through a stochastic differential equation (SDE) that governs the collective dynamics of the group. We apply this technique on both synthetic and real-world datasets, and identify the deterministic and stochastic aspects of the dynamics using drift and diffusion fields, enabling us to make novel inferences about the nature of order in these systems.
ALM-KD: Knowledge Distillation with noisy labels via adaptive loss mixing
Feb 07, 2022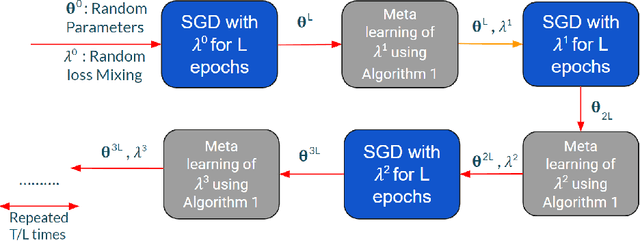
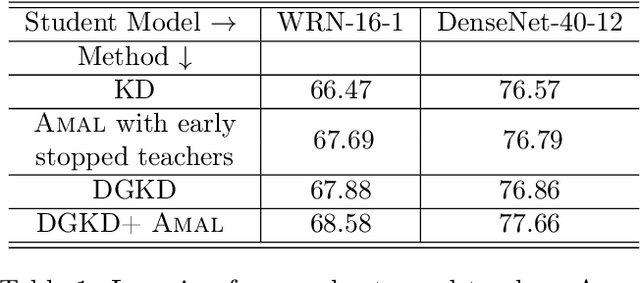
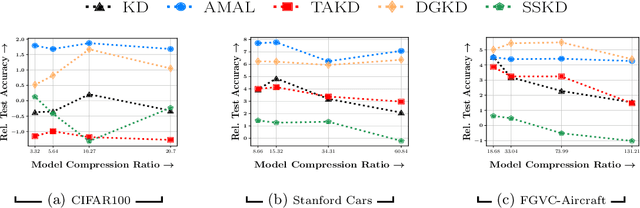
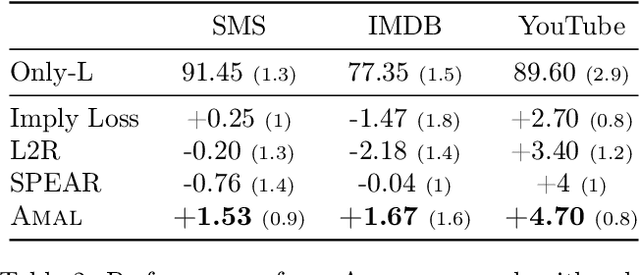
Abstract:Knowledge distillation is a technique where the outputs of a pretrained model, often known as the teacher model is used for training a student model in a supervised setting. The teacher model outputs being a richer distribution over labels should improve the student model's performance as opposed to training with the usual hard labels. However, the label distribution imposed by the logits of the teacher network may not be always informative and may lead to poor student performance. We tackle this problem via the use of an adaptive loss mixing scheme during KD. Specifically, our method learns an instance-specific convex combination of the teacher-matching and label supervision objectives, using meta learning on a validation metric signalling to the student `how much' of KD is to be used. Through a range of experiments on controlled synthetic data and real-world datasets, we demonstrate performance gains obtained using our approach in the standard KD setting as well as in multi-teacher and self-distillation settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge