Patrick Schwab
CausalBench: A Large-scale Benchmark for Network Inference from Single-cell Perturbation Data
Oct 31, 2022Abstract:Mapping biological mechanisms in cellular systems is a fundamental step in early-stage drug discovery that serves to generate hypotheses on what disease-relevant molecular targets may effectively be modulated by pharmacological interventions. With the advent of high-throughput methods for measuring single-cell gene expression under genetic perturbations, we now have effective means for generating evidence for causal gene-gene interactions at scale. However, inferring graphical networks of the size typically encountered in real-world gene-gene interaction networks is difficult in terms of both achieving and evaluating faithfulness to the true underlying causal graph. Moreover, standardised benchmarks for comparing methods for causal discovery in perturbational single-cell data do not yet exist. Here, we introduce CausalBench - a comprehensive benchmark suite for evaluating network inference methods on large-scale perturbational single-cell gene expression data. CausalBench introduces several biologically meaningful performance metrics and operates on two large, curated and openly available benchmark data sets for evaluating methods on the inference of gene regulatory networks from single-cell data generated under perturbations. With real-world datasets consisting of over \numprint{200000} training samples under interventions, CausalBench could potentially help facilitate advances in causal network inference by providing what is - to the best of our knowledge - the largest openly available test bed for causal discovery from real-world perturbation data to date.
Federated Learning in Multi-Center Critical Care Research: A Systematic Case Study using the eICU Database
Apr 20, 2022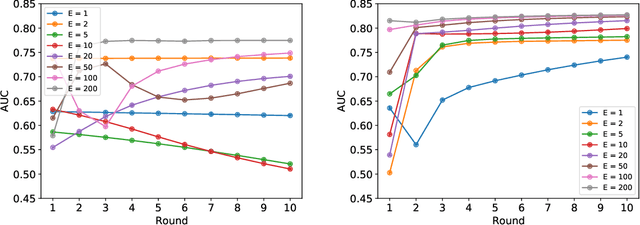
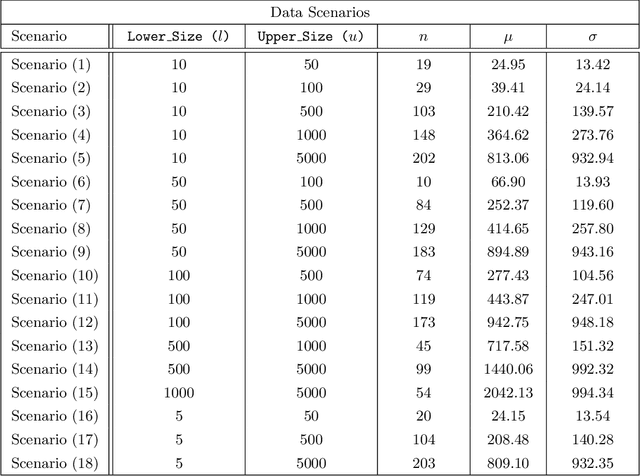
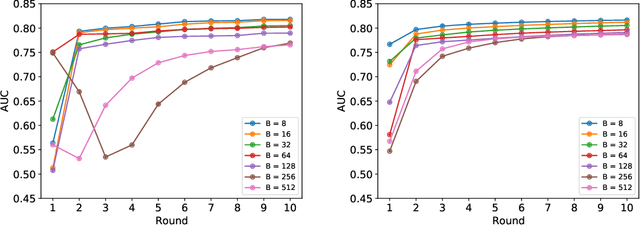
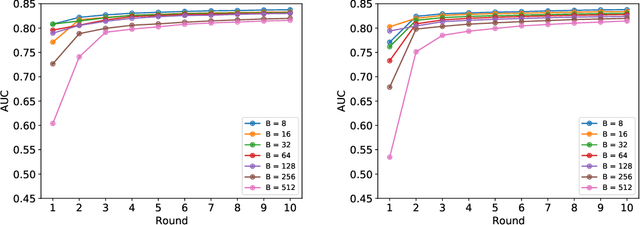
Abstract:Federated learning (FL) has been proposed as a method to train a model on different units without exchanging data. This offers great opportunities in the healthcare sector, where large datasets are available but cannot be shared to ensure patient privacy. We systematically investigate the effectiveness of FL on the publicly available eICU dataset for predicting the survival of each ICU stay. We employ Federated Averaging as the main practical algorithm for FL and show how its performance changes by altering three key hyper-parameters, taking into account that clients can significantly vary in size. We find that in many settings, a large number of local training epochs improves the performance while at the same time reducing communication costs. Furthermore, we outline in which settings it is possible to have only a low number of hospitals participating in each federated update round. When many hospitals with low patient counts are involved, the effect of overfitting can be avoided by decreasing the batchsize. This study thus contributes toward identifying suitable settings for running distributed algorithms such as FL on clinical datasets.
Conditional Generation of Medical Time Series for Extrapolation to Underrepresented Populations
Jan 20, 2022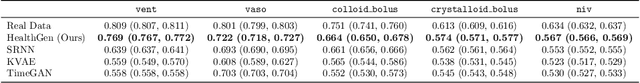
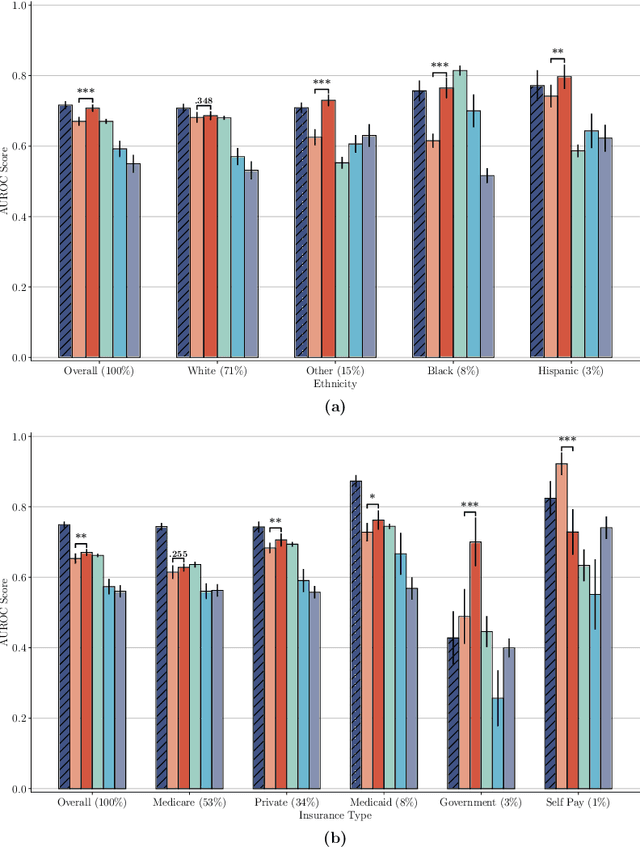
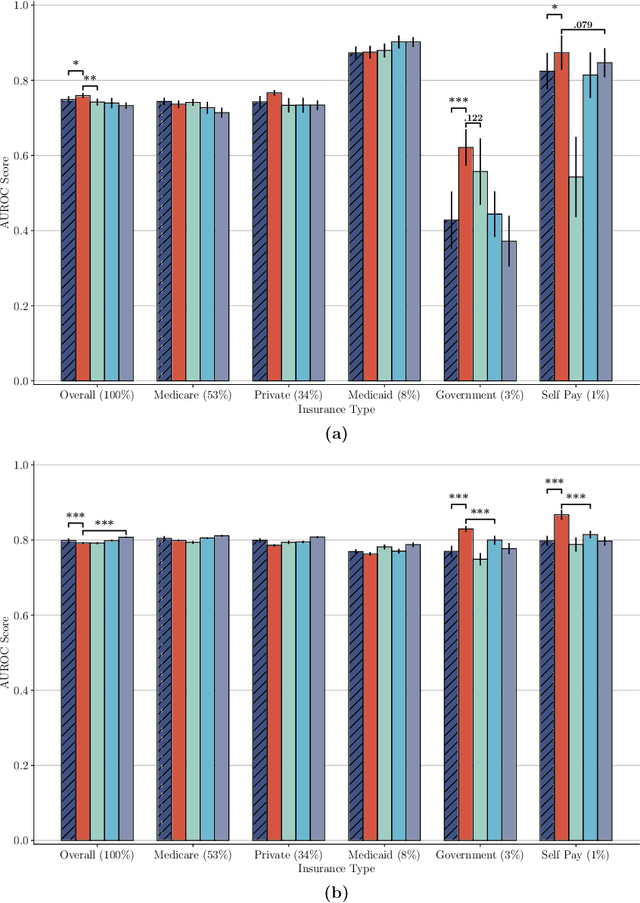
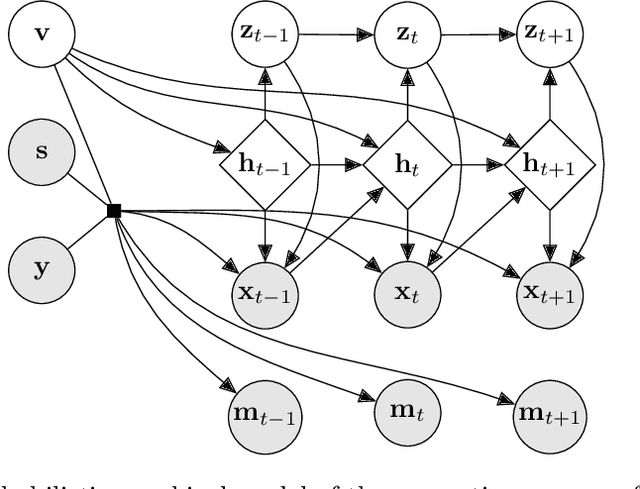
Abstract:The widespread adoption of electronic health records (EHRs) and subsequent increased availability of longitudinal healthcare data has led to significant advances in our understanding of health and disease with direct and immediate impact on the development of new diagnostics and therapeutic treatment options. However, access to EHRs is often restricted due to their perceived sensitive nature and associated legal concerns, and the cohorts therein typically are those seen at a specific hospital or network of hospitals and therefore not representative of the wider population of patients. Here, we present HealthGen, a new approach for the conditional generation of synthetic EHRs that maintains an accurate representation of real patient characteristics, temporal information and missingness patterns. We demonstrate experimentally that HealthGen generates synthetic cohorts that are significantly more faithful to real patient EHRs than the current state-of-the-art, and that augmenting real data sets with conditionally generated cohorts of underrepresented subpopulations of patients can significantly enhance the generalisability of models derived from these data sets to different patient populations. Synthetic conditionally generated EHRs could help increase the accessibility of longitudinal healthcare data sets and improve the generalisability of inferences made from these data sets to underrepresented populations.
GeneDisco: A Benchmark for Experimental Design in Drug Discovery
Oct 22, 2021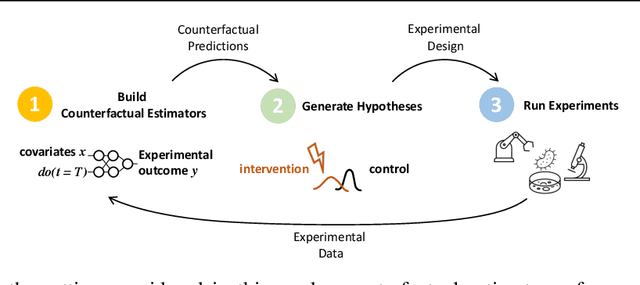
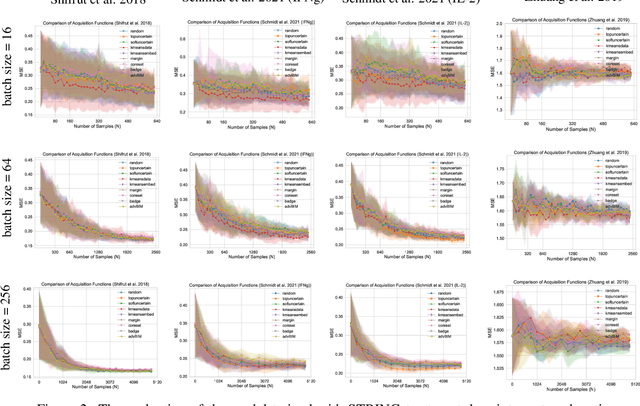
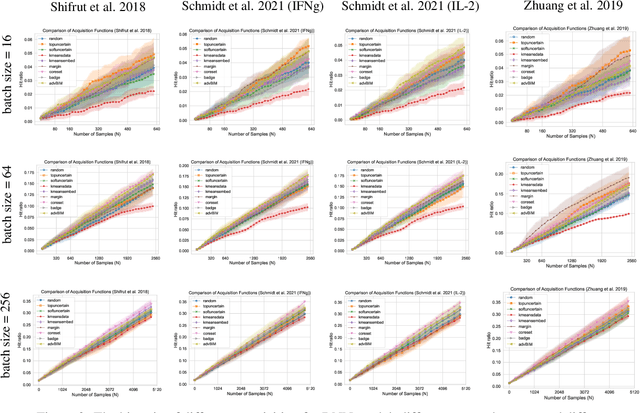
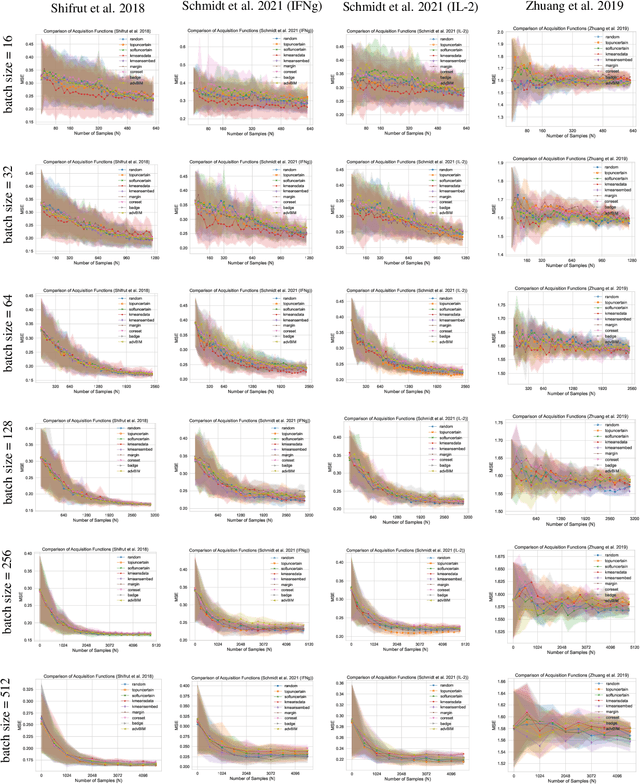
Abstract:In vitro cellular experimentation with genetic interventions, using for example CRISPR technologies, is an essential step in early-stage drug discovery and target validation that serves to assess initial hypotheses about causal associations between biological mechanisms and disease pathologies. With billions of potential hypotheses to test, the experimental design space for in vitro genetic experiments is extremely vast, and the available experimental capacity - even at the largest research institutions in the world - pales in relation to the size of this biological hypothesis space. Machine learning methods, such as active and reinforcement learning, could aid in optimally exploring the vast biological space by integrating prior knowledge from various information sources as well as extrapolating to yet unexplored areas of the experimental design space based on available data. However, there exist no standardised benchmarks and data sets for this challenging task and little research has been conducted in this area to date. Here, we introduce GeneDisco, a benchmark suite for evaluating active learning algorithms for experimental design in drug discovery. GeneDisco contains a curated set of multiple publicly available experimental data sets as well as open-source implementations of state-of-the-art active learning policies for experimental design and exploration.
Learning Neural Causal Models with Active Interventions
Sep 06, 2021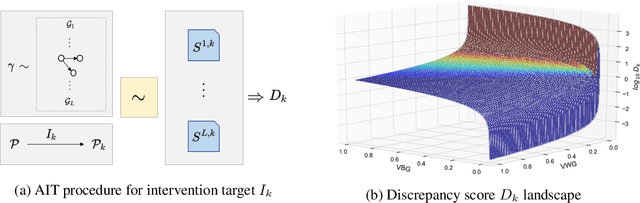
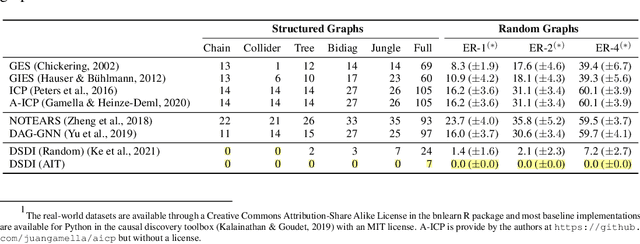
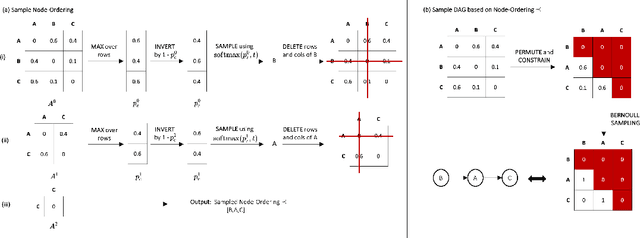
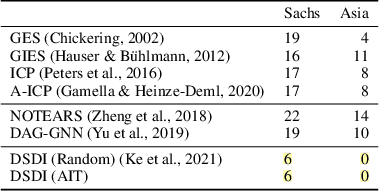
Abstract:Discovering causal structures from data is a challenging inference problem of fundamental importance in all areas of science. The appealing scaling properties of neural networks have recently led to a surge of interest in differentiable neural network-based methods for learning causal structures from data. So far differentiable causal discovery has focused on static datasets of observational or interventional origin. In this work, we introduce an active intervention-targeting mechanism which enables a quick identification of the underlying causal structure of the data-generating process. Our method significantly reduces the required number of interactions compared with random intervention targeting and is applicable for both discrete and continuous optimization formulations of learning the underlying directed acyclic graph (DAG) from data. We examine the proposed method across a wide range of settings and demonstrate superior performance on multiple benchmarks from simulated to real-world data.
NCoRE: Neural Counterfactual Representation Learning for Combinations of Treatments
Mar 20, 2021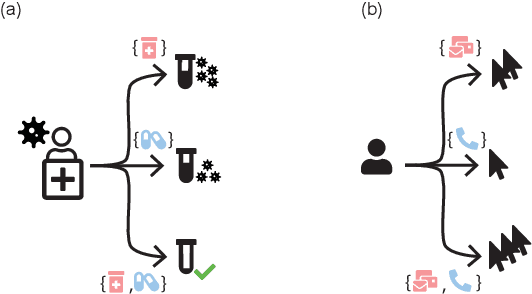
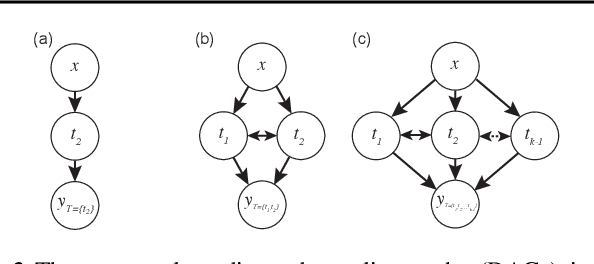
Abstract:Estimating an individual's potential response to interventions from observational data is of high practical relevance for many domains, such as healthcare, public policy or economics. In this setting, it is often the case that combinations of interventions may be applied simultaneously, for example, multiple prescriptions in healthcare or different fiscal and monetary measures in economics. However, existing methods for counterfactual inference are limited to settings in which actions are not used simultaneously. Here, we present Neural Counterfactual Relation Estimation (NCoRE), a new method for learning counterfactual representations in the combination treatment setting that explicitly models cross-treatment interactions. NCoRE is based on a novel branched conditional neural representation that includes learnt treatment interaction modulators to infer the potential causal generative process underlying the combination of multiple treatments. Our experiments show that NCoRE significantly outperforms existing state-of-the-art methods for counterfactual treatment effect estimation that do not account for the effects of combining multiple treatments across several synthetic, semi-synthetic and real-world benchmarks.
Overcoming Barriers to Data Sharing with Medical Image Generation: A Comprehensive Evaluation
Nov 29, 2020
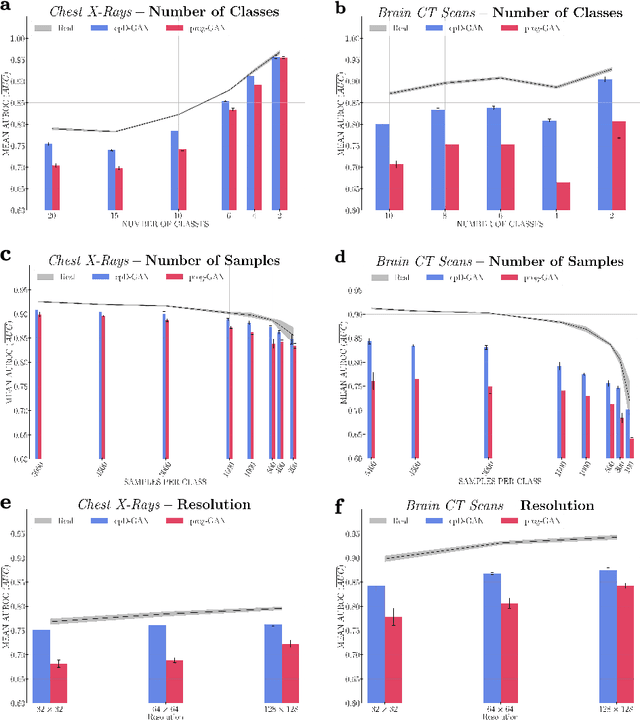
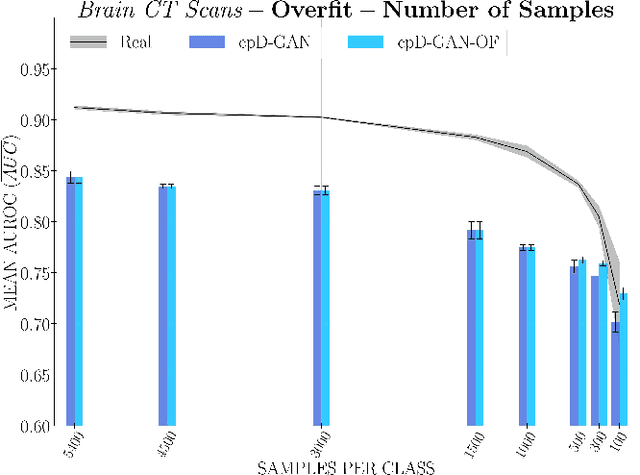
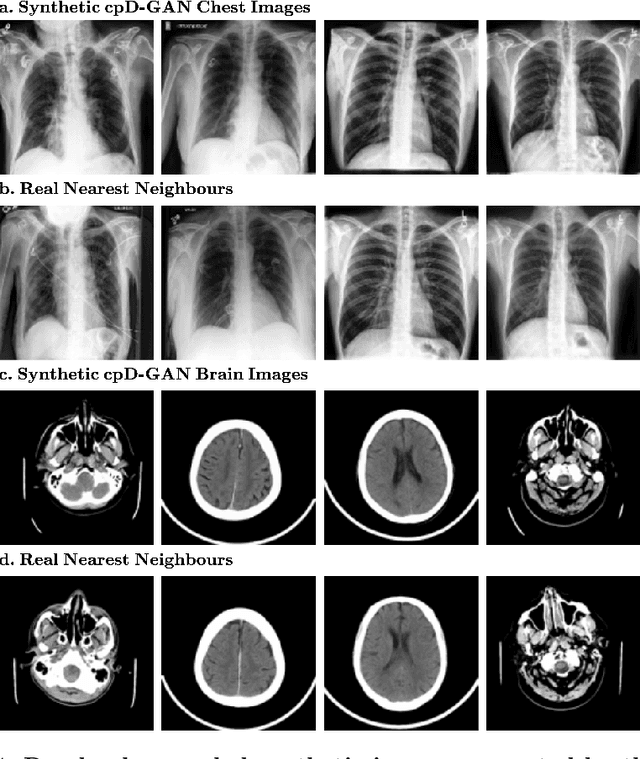
Abstract:Privacy concerns around sharing personally identifiable information are a major practical barrier to data sharing in medical research. However, in many cases, researchers have no interest in a particular individual's information but rather aim to derive insights at the level of cohorts. Here, we utilize Generative Adversarial Networks (GANs) to create derived medical imaging datasets consisting entirely of synthetic patient data. The synthetic images ideally have, in aggregate, similar statistical properties to those of a source dataset but do not contain sensitive personal information. We assess the quality of synthetic data generated by two GAN models for chest radiographs with 14 different radiology findings and brain computed tomography (CT) scans with six types of intracranial hemorrhages. We measure the synthetic image quality by the performance difference of predictive models trained on either the synthetic or the real dataset. We find that synthetic data performance disproportionately benefits from a reduced number of unique label combinations and determine at what number of samples per class overfitting effects start to dominate GAN training. Our open-source benchmark findings also indicate that synthetic data generation can benefit from higher levels of spatial resolution. We additionally conducted a reader study in which trained radiologists do not perform better than random on discriminating between synthetic and real medical images for both data modalities to a statistically significant extent. Our study offers valuable guidelines and outlines practical conditions under which insights derived from synthetic medical images are similar to those that would have been derived from real imaging data. Our results indicate that synthetic data sharing may be an attractive and privacy-preserving alternative to sharing real patient-level data in the right settings.
Real-time Prediction of COVID-19 related Mortality using Electronic Health Records
Aug 31, 2020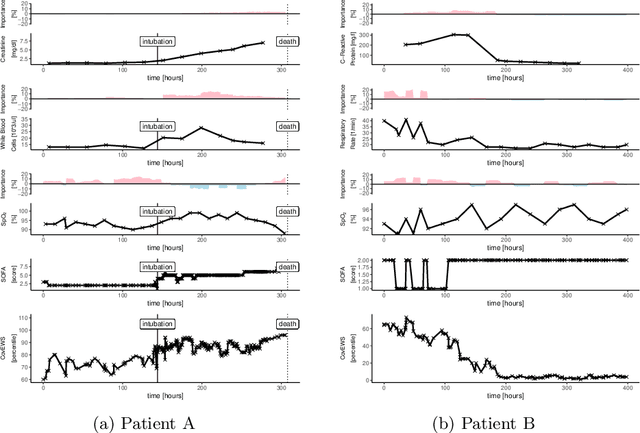
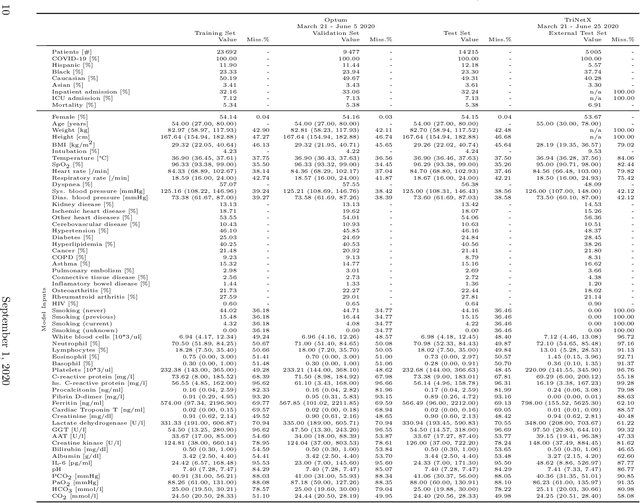
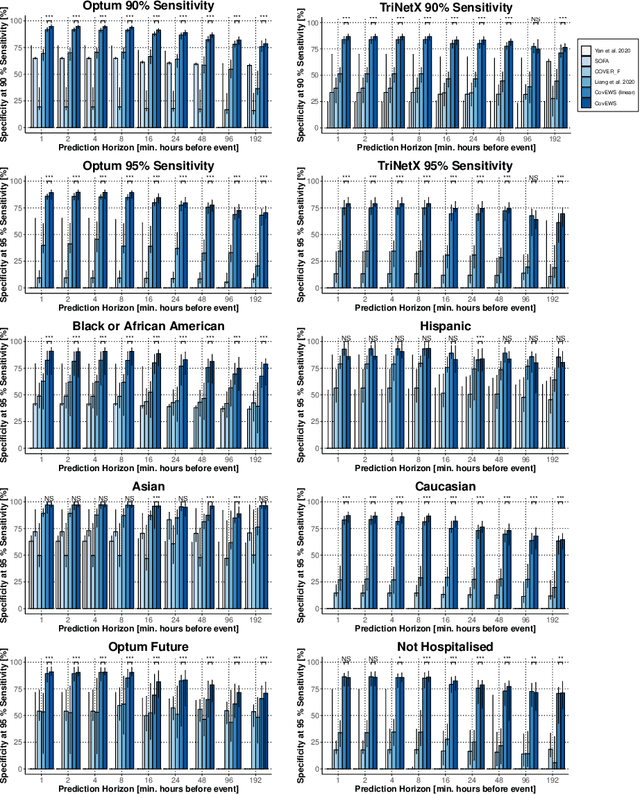
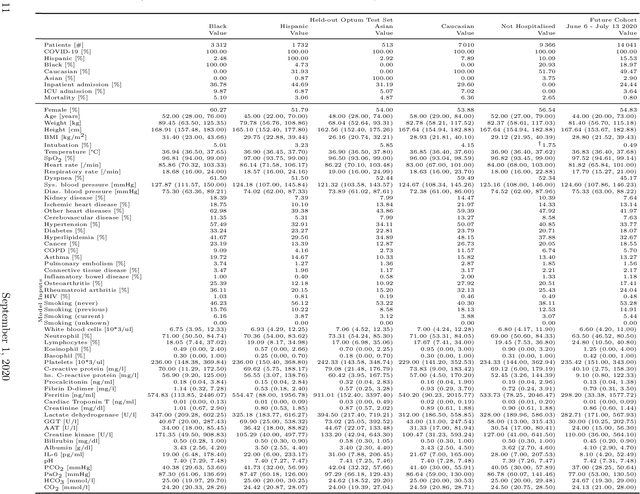
Abstract:Coronavirus Disease 2019 (COVID-19) is an emerging respiratory disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with rapid human-to-human transmission and a high case fatality rate particularly in older patients. Due to the exponential growth of infections, many healthcare systems across the world are under pressure to care for increasing amounts of at-risk patients. Given the high number of infected patients, identifying patients with the highest mortality risk early is critical to enable effective intervention and optimal prioritisation of care. Here, we present the COVID-19 Early Warning System (CovEWS), a clinical risk scoring system for assessing COVID-19 related mortality risk. CovEWS provides continuous real-time risk scores for individual patients with clinically meaningful predictive performance up to 192 hours (8 days) in advance, and is automatically derived from patients' electronic health records (EHRs) using machine learning. We trained and evaluated CovEWS using de-identified data from a cohort of 66430 COVID-19 positive patients seen at over 69 healthcare institutions in the United States (US), Australia, Malaysia and India amounting to an aggregated total of over 2863 years of patient observation time. On an external test cohort of 5005 patients, CovEWS predicts COVID-19 related mortality from $78.8\%$ ($95\%$ confidence interval [CI]: $76.0$, $84.7\%$) to $69.4\%$ ($95\%$ CI: $57.6, 75.2\%$) specificity at a sensitivity greater than $95\%$ between respectively 1 and 192 hours prior to observed mortality events - significantly outperforming existing generic and COVID-19 specific clinical risk scores. CovEWS could enable clinicians to intervene at an earlier stage, and may therefore help in preventing or mitigating COVID-19 related mortality.
predCOVID-19: A Systematic Study of Clinical Predictive Models for Coronavirus Disease 2019
May 17, 2020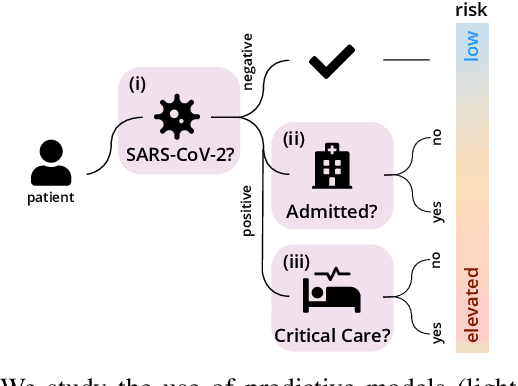
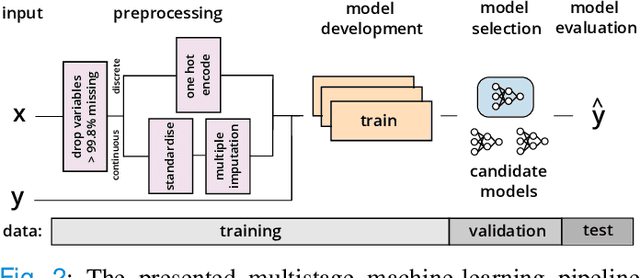
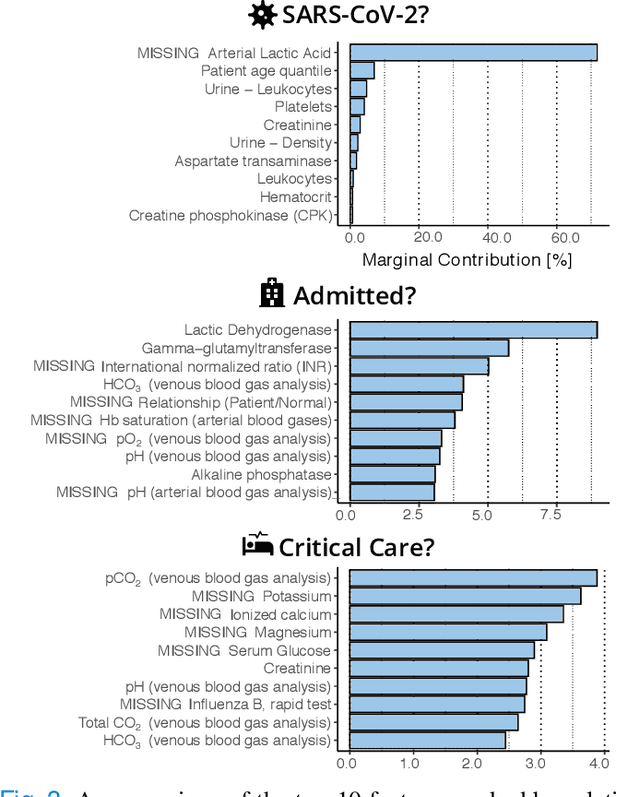
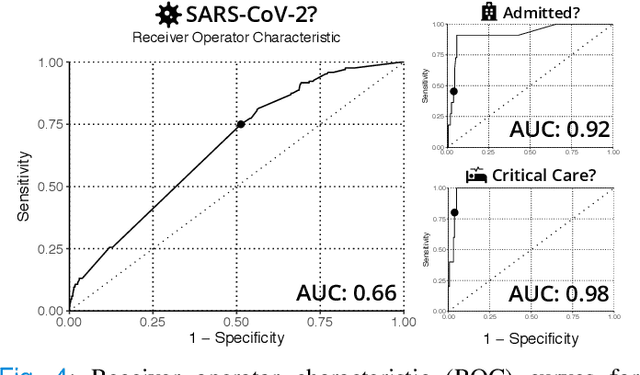
Abstract:Coronavirus Disease 2019 (COVID-19) is a rapidly emerging respiratory disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Due to the rapid human-to-human transmission of SARS-CoV-2, many healthcare systems are at risk of exceeding their healthcare capacities, in particular in terms of SARS-CoV-2 tests, hospital and intensive care unit (ICU) beds and mechanical ventilators. Predictive algorithms could potentially ease the strain on healthcare systems by identifying those who are most likely to receive a positive SARS-CoV-2 test, be hospitalised or admitted to the ICU. Here, we study clinical predictive models that estimate, using machine learning and based on routinely collected clinical data, which patients are likely to receive a positive SARS-CoV-2 test, require hospitalisation or intensive care. To evaluate the predictive performance of our models, we perform a retrospective evaluation on clinical and blood analysis data from a cohort of 5644 patients. Our experimental results indicate that our predictive models identify (i) patients that test positive for SARS-CoV-2 a priori at a sensitivity of 75% (95% CI: 67%, 81%) and a specificity of 49% (95% CI: 46%, 51%), (ii) SARS-CoV-2 positive patients that require hospitalisation with 0.92 AUC (95% CI: 0.81, 0.98), and (iii) SARS-CoV-2 positive patients that require critical care with 0.98 AUC (95% CI: 0.95, 1.00). In addition, we determine which clinical features are predictive to what degree for each of the aforementioned clinical tasks. Our results indicate that predictive models trained on routinely collected clinical data could be used to predict clinical pathways for COVID-19, and therefore help inform care and prioritise resources.
A Deep Learning Approach to Diagnosing Multiple Sclerosis from Smartphone Data
Jan 02, 2020
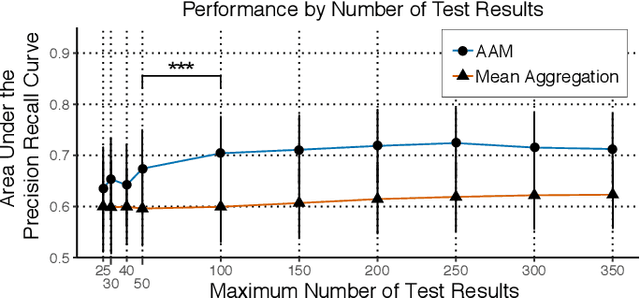
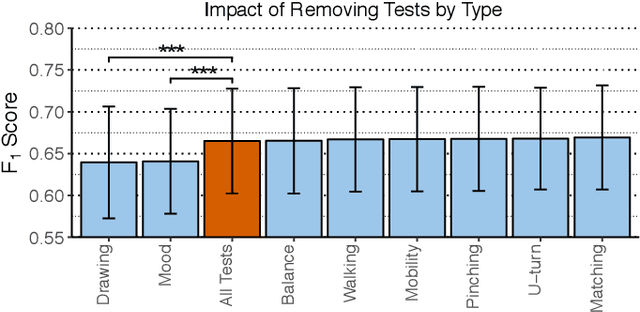
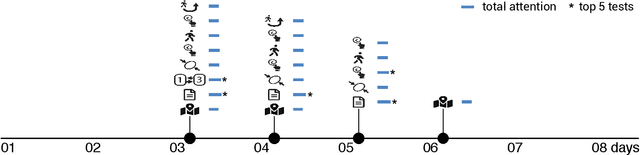
Abstract:Multiple sclerosis (MS) affects the central nervous system with a wide range of symptoms. MS can, for example, cause pain, changes in mood and fatigue, and may impair a person's movement, speech and visual functions. Diagnosis of MS typically involves a combination of complex clinical assessments and tests to rule out other diseases with similar symptoms. New technologies, such as smartphone monitoring in free-living conditions, could potentially aid in objectively assessing the symptoms of MS by quantifying symptom presence and intensity over long periods of time. Here, we present a deep-learning approach to diagnosing MS from smartphone-derived digital biomarkers that uses a novel combination of a multilayer perceptron with neural soft attention to improve learning of patterns in long-term smartphone monitoring data. Using data from a cohort of 774 participants, we demonstrate that our deep-learning models are able to distinguish between people with and without MS with an area under the receiver operating characteristic curve of 0.88 (95% CI: 0.70, 0.88). Our experimental results indicate that digital biomarkers derived from smartphone data could in the future be used as additional diagnostic criteria for MS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge