Thomas A. W. Bolton
Department of Radiology, Lausanne University Hospital and University of Lausanne
Comparing and Scaling fMRI Features for Brain-Behavior Prediction
Jul 28, 2025Abstract:Predicting behavioral variables from neuroimaging modalities such as magnetic resonance imaging (MRI) has the potential to allow the development of neuroimaging biomarkers of mental and neurological disorders. A crucial processing step to this aim is the extraction of suitable features. These can differ in how well they predict the target of interest, and how this prediction scales with sample size and scan time. Here, we compare nine feature subtypes extracted from resting-state functional MRI recordings for behavior prediction, ranging from regional measures of functional activity to functional connectivity (FC) and metrics derived with graph signal processing (GSP), a principled approach for the extraction of structure-informed functional features. We study 979 subjects from the Human Connectome Project Young Adult dataset, predicting summary scores for mental health, cognition, processing speed, and substance use, as well as age and sex. The scaling properties of the features are investigated for different combinations of sample size and scan time. FC comes out as the best feature for predicting cognition, age, and sex. Graph power spectral density is the second best for predicting cognition and age, while for sex, variability-based features show potential as well. When predicting sex, the low-pass graph filtered coupled FC slightly outperforms the simple FC variant. None of the other targets were predicted significantly. The scaling results point to higher performance reserves for the better-performing features. They also indicate that it is important to balance sample size and scan time when acquiring data for prediction studies. The results confirm FC as a robust feature for behavior prediction, but also show the potential of GSP and variability-based measures. We discuss the implications for future prediction studies in terms of strategies for acquisition and sample composition.
Predicting Cognition from fMRI:A Comparative Study of Graph, Transformer, and Kernel Models Across Task and Rest Conditions
Jul 28, 2025Abstract:Predicting cognition from neuroimaging data in healthy individuals offers insights into the neural mechanisms underlying cognitive abilities, with potential applications in precision medicine and early detection of neurological and psychiatric conditions. This study systematically benchmarked classical machine learning (Kernel Ridge Regression (KRR)) and advanced deep learning (DL) models (Graph Neural Networks (GNN) and Transformer-GNN (TGNN)) for cognitive prediction using Resting-state (RS), Working Memory, and Language task fMRI data from the Human Connectome Project Young Adult dataset. Our results, based on R2 scores, Pearson correlation coefficient, and mean absolute error, revealed that task-based fMRI, eliciting neural responses directly tied to cognition, outperformed RS fMRI in predicting cognitive behavior. Among the methods compared, a GNN combining structural connectivity (SC) and functional connectivity (FC) consistently achieved the highest performance across all fMRI modalities; however, its advantage over KRR using FC alone was not statistically significant. The TGNN, designed to model temporal dynamics with SC as a prior, performed competitively with FC-based approaches for task-fMRI but struggled with RS data, where its performance aligned with the lower-performing GNN that directly used fMRI time-series data as node features. These findings emphasize the importance of selecting appropriate model architectures and feature representations to fully leverage the spatial and temporal richness of neuroimaging data. This study highlights the potential of multimodal graph-aware DL models to combine SC and FC for cognitive prediction, as well as the promise of Transformer-based approaches for capturing temporal dynamics. By providing a comprehensive comparison of models, this work serves as a guide for advancing brain-behavior modeling using fMRI, SC and DL.
Guided Graph Spectral Embedding: Application to the C. elegans Connectome
Dec 10, 2018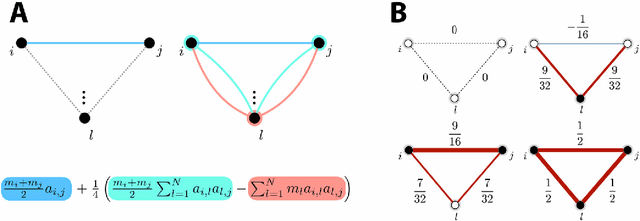
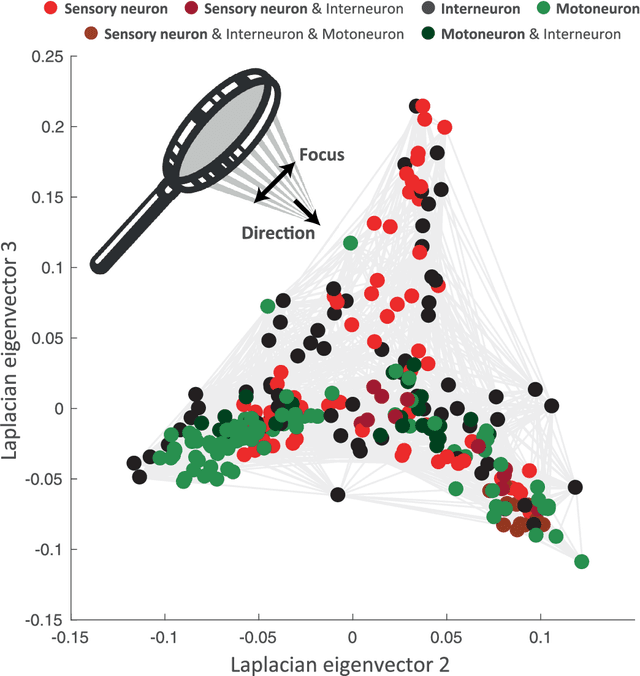
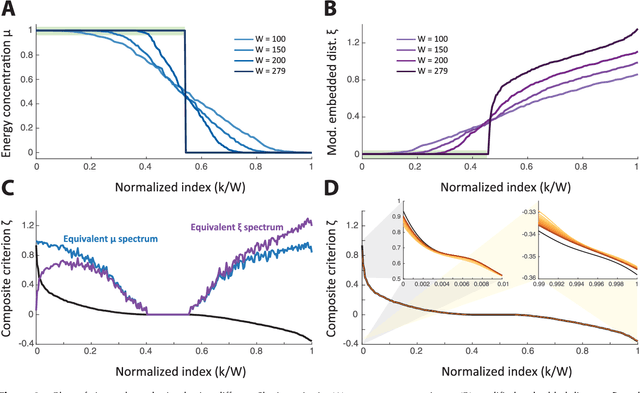
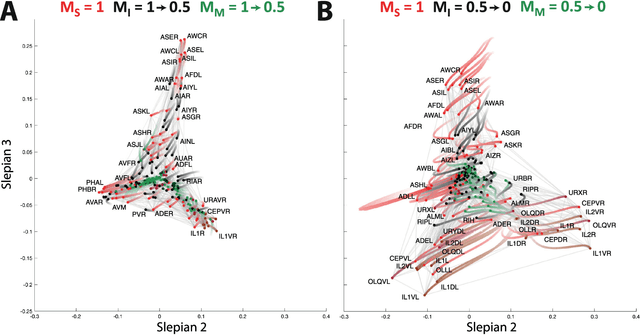
Abstract:Graph spectral analysis can yield meaningful embeddings of graphs by providing insight into distributed features not directly accessible in nodal domain. Recent efforts in graph signal processing have proposed new decompositions-e.g., based on wavelets and Slepians-that can be applied to filter signals defined on the graph. In this work, we take inspiration from these constructions to define a new guided spectral embedding that combines maximizing energy concentration with minimizing modified embedded distance for a given importance weighting of the nodes. We show these optimization goals are intrinsically opposite, leading to a well-defined and stable spectral decomposition. The importance weighting allows to put the focus on particular nodes and tune the trade-off between global and local effects. Following the derivation of our new optimization criterion and its linear approximation, we exemplify the methodology on the C. elegans structural connectome. The results of our analyses confirm known observations on the nematode's neural network in terms of functionality and importance of cells. Compared to Laplacian embedding, the guided approach, focused on a certain class of cells (sensory, inter- and motoneurons), provides more biological insights, such as the distinction between somatic positions of cells, and their involvement in low or high order processing functions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge