Oliver Hayden
Reconstructing Patched or Partial Holograms to allow for Whole Slide Imaging with a Self-Referencing Holographic Microscope
Jan 22, 2026Abstract:The last decade has seen significant advances in computer-aided diagnostics for cytological screening, mainly through the improvement and integration of scanning techniques such as whole slide imaging (WSI) and the combination with deep learning. Simultaneously, new imaging techniques such as quantitative phase imaging (QPI) are being developed to capture richer cell information with less sample preparation. So far, the two worlds of WSI and QPI have not been combined. In this work, we present a reconstruction algorithm which makes whole slide imaging of cervical smears possible by using a self-referencing three-wave digital holographic microscope. Since a WSI is constructed by combining multiple patches, the algorithm is adaptive and can be used on partial holograms and patched holograms. We present the algorithm for a single shot hologram, the adaptations to make it flexible to various inputs and show that the algorithm performs well for the tested epithelial cells. This is a preprint of our paper, which has been accepted for publication in 2026 IEEE International Symposium on Biomedical Imaging (ISBI).
Disentangling coincident cell events using deep transfer learning and compressive sensing
Jul 17, 2025Abstract:Accurate single-cell analysis is critical for diagnostics, immunomonitoring, and cell therapy, but coincident events - where multiple cells overlap in a sensing zone - can severely compromise signal fidelity. We present a hybrid framework combining a fully convolutional neural network (FCN) with compressive sensing (CS) to disentangle such overlapping events in one-dimensional sensor data. The FCN, trained on bead-derived datasets, accurately estimates coincident event counts and generalizes to immunomagnetically labeled CD4+ and CD14+ cells in whole blood without retraining. Using this count, the CS module reconstructs individual signal components with high fidelity, enabling precise recovery of single-cell features, including velocity, amplitude, and hydrodynamic diameter. Benchmarking against conventional state-machine algorithms shows superior performance - recovering up to 21% more events and improving classification accuracy beyond 97%. Explinability via class activation maps and parameterized Gaussian template fitting ensures transparency and clinical interpretability. Demonstrated with magnetic flow cytometry (MFC), the framework is compatible with other waveform-generating modalities, including impedance cytometry, nanopore, and resistive pulse sensing. This work lays the foundation for next-generation non-optical single-cell sensing platforms that are automated, generalizable, and capable of resolving overlapping events, broadening the utility of cytometry in translational medicine and precision diagnostics, e.g. cell-interaction studies.
OAH-Net: A Deep Neural Network for Hologram Reconstruction of Off-axis Digital Holographic Microscope
Oct 17, 2024



Abstract:Off-axis digital holographic microscopy is a high-throughput, label-free imaging technology that provides three-dimensional, high-resolution information about samples, particularly useful in large-scale cellular imaging. However, the hologram reconstruction process poses a significant bottleneck for timely data analysis. To address this challenge, we propose a novel reconstruction approach that integrates deep learning with the physical principles of off-axis holography. We initialized part of the network weights based on the physical principle and then fine-tuned them via weakly supersized learning. Our off-axis hologram network (OAH-Net) retrieves phase and amplitude images with errors that fall within the measurement error range attributable to hardware, and its reconstruction speed significantly surpasses the microscope's acquisition rate. Crucially, OAH-Net demonstrates remarkable external generalization capabilities on unseen samples with distinct patterns and can be seamlessly integrated with other models for downstream tasks to achieve end-to-end real-time hologram analysis. This capability further expands off-axis holography's applications in both biological and medical studies.
Unsupervised high-throughput segmentation of cells and cell nuclei in quantitative phase images
Nov 24, 2023



Abstract:In the effort to aid cytologic diagnostics by establishing automatic single cell screening using high throughput digital holographic microscopy for clinical studies thousands of images and millions of cells are captured. The bottleneck lies in an automatic, fast, and unsupervised segmentation technique that does not limit the types of cells which might occur. We propose an unsupervised multistage method that segments correctly without confusing noise or reflections with cells and without missing cells that also includes the detection of relevant inner structures, especially the cell nucleus in the unstained cell. In an effort to make the information reasonable and interpretable for cytopathologists, we also introduce new cytoplasmic and nuclear features of potential help for cytologic diagnoses which exploit the quantitative phase information inherent to the measurement scheme. We show that the segmentation provides consistently good results over many experiments on patient samples in a reasonable per cell analysis time.
Towards Interpretable Classification of Leukocytes based on Deep Learning
Nov 24, 2023



Abstract:Label-free approaches are attractive in cytological imaging due to their flexibility and cost efficiency. They are supported by machine learning methods, which, despite the lack of labeling and the associated lower contrast, can classify cells with high accuracy where the human observer has little chance to discriminate cells. In order to better integrate these workflows into the clinical decision making process, this work investigates the calibration of confidence estimation for the automated classification of leukocytes. In addition, different visual explanation approaches are compared, which should bring machine decision making closer to professional healthcare applications. Furthermore, we were able to identify general detection patterns in neural networks and demonstrate the utility of the presented approaches in different scenarios of blood cell analysis.
Outlier Detection using Self-Organizing Maps for Automated Blood Cell Analysis
Aug 18, 2022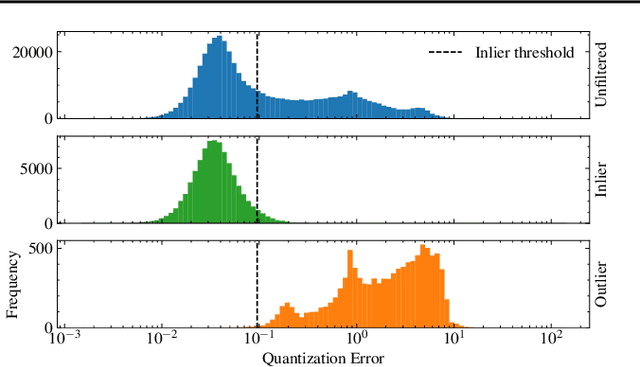
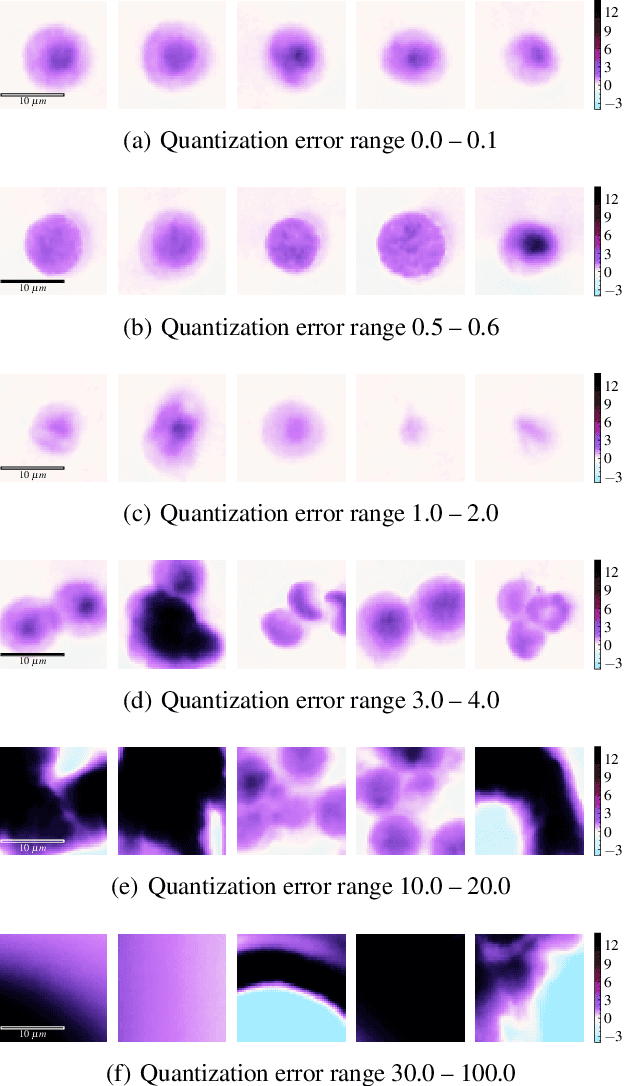
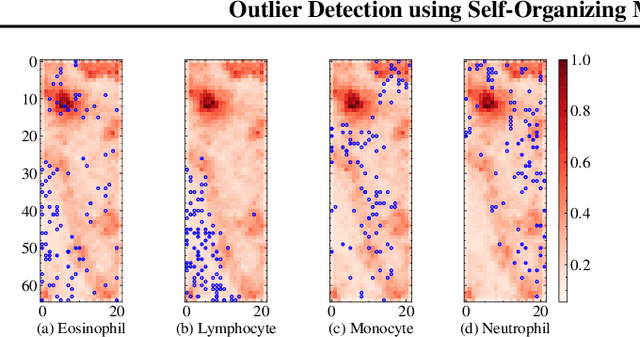
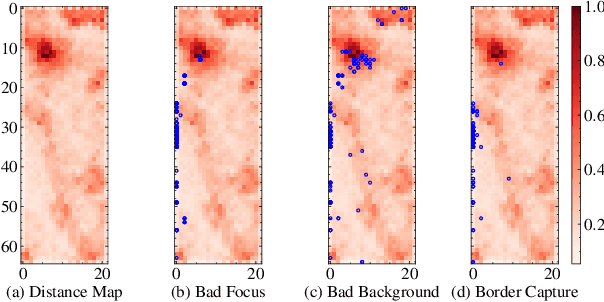
Abstract:The quality of datasets plays a crucial role in the successful training and deployment of deep learning models. Especially in the medical field, where system performance may impact the health of patients, clean datasets are a safety requirement for reliable predictions. Therefore, outlier detection is an essential process when building autonomous clinical decision systems. In this work, we assess the suitability of Self-Organizing Maps for outlier detection specifically on a medical dataset containing quantitative phase images of white blood cells. We detect and evaluate outliers based on quantization errors and distance maps. Our findings confirm the suitability of Self-Organizing Maps for unsupervised Out-Of-Distribution detection on the dataset at hand. Self-Organizing Maps perform on par with a manually specified filter based on expert domain knowledge. Additionally, they show promise as a tool in the exploration and cleaning of medical datasets. As a direction for future research, we suggest a combination of Self-Organizing Maps and feature extraction based on deep learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge