Nithya Bhasker
Adaptive-CaRe: Adaptive Causal Regularization for Robust Outcome Prediction
Feb 06, 2026Abstract:Accurate prediction of outcomes is crucial for clinical decision-making and personalized patient care. Supervised machine learning algorithms, which are commonly used for outcome prediction in the medical domain, optimize for predictive accuracy, which can result in models latching onto spurious correlations instead of robust predictors. Causal structure learning methods on the other hand have the potential to provide robust predictors for the target, but can be too conservative because of algorithmic and data assumptions, resulting in loss of diagnostic precision. Therefore, we propose a novel model-agnostic regularization strategy, Adaptive-CaRe, for generalized outcome prediction in the medical domain. Adaptive-CaRe strikes a balance between both predictive value and causal robustness by incorporating a penalty that is proportional to the difference between the estimated statistical contribution and estimated causal contribution of the input features for model predictions. Our experiments on synthetic data establish the efficacy of the proposed Adaptive-CaRe regularizer in finding robust predictors for the target while maintaining competitive predictive accuracy. With experiments on a standard causal benchmark, we provide a blueprint for navigating the trade-off between predictive accuracy and causal robustness by tweaking the regularization strength, $λ$. Validation using real-world dataset confirms that the results translate to practical, real-domain settings. Therefore, Adaptive-CaRe provides a simple yet effective solution to the long-standing trade-off between predictive accuracy and causal robustness in the medical domain. Future work would involve studying alternate causal structure learning frameworks and complex classification models to provide deeper insights at a larger scale.
HistoPrism: Unlocking Functional Pathway Analysis from Pan-Cancer Histology via Gene Expression Prediction
Jan 29, 2026Abstract:Predicting spatial gene expression from H&E histology offers a scalable and clinically accessible alternative to sequencing, but realizing clinical impact requires models that generalize across cancer types and capture biologically coherent signals. Prior work is often limited to per-cancer settings and variance-based evaluation, leaving functional relevance underexplored. We introduce HistoPrism, an efficient transformer-based architecture for pan-cancer prediction of gene expression from histology. To evaluate biological meaning, we introduce a pathway-level benchmark, shifting assessment from isolated gene-level variance to coherent functional pathways. HistoPrism not only surpasses prior state-of-the-art models on highly variable genes , but also more importantly, achieves substantial gains on pathway-level prediction, demonstrating its ability to recover biologically coherent transcriptomic patterns. With strong pan-cancer generalization and improved efficiency, HistoPrism establishes a new standard for clinically relevant transcriptomic modeling from routinely available histology.
* Accepted at ICLR2026
Graph data modelling for outcome prediction in oropharyngeal cancer patients
Oct 04, 2023



Abstract:Graph neural networks (GNNs) are becoming increasingly popular in the medical domain for the tasks of disease classification and outcome prediction. Since patient data is not readily available as a graph, most existing methods either manually define a patient graph, or learn a latent graph based on pairwise similarities between the patients. There are also hypergraph neural network (HGNN)-based methods that were introduced recently to exploit potential higher order associations between the patients by representing them as a hypergraph. In this work, we propose a patient hypergraph network (PHGN), which has been investigated in an inductive learning setup for binary outcome prediction in oropharyngeal cancer (OPC) patients using computed tomography (CT)-based radiomic features for the first time. Additionally, the proposed model was extended to perform time-to-event analyses, and compared with GNN and baseline linear models.
CholecTriplet2021: A benchmark challenge for surgical action triplet recognition
Apr 10, 2022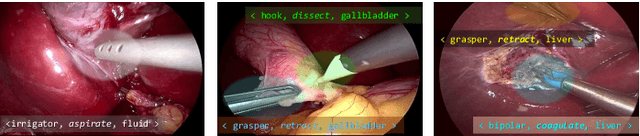
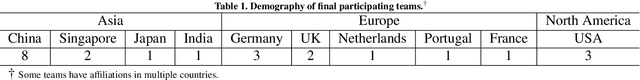
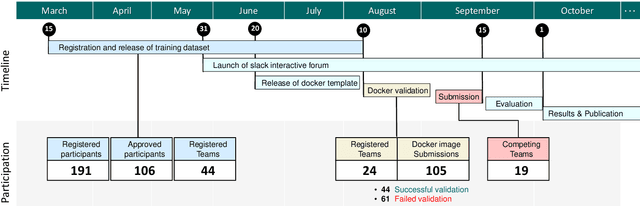
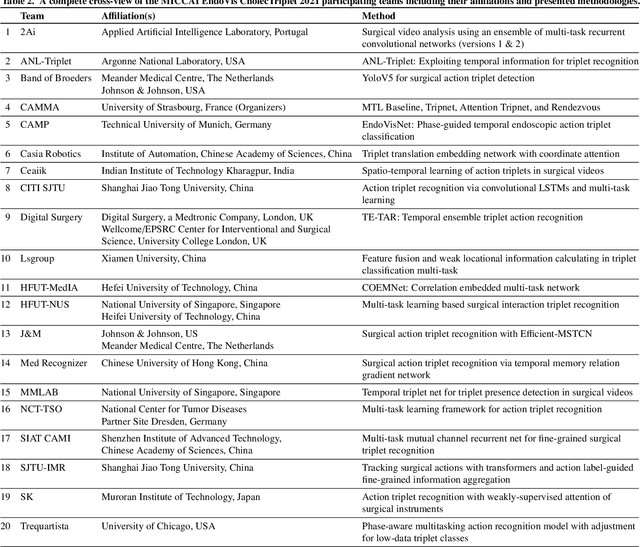
Abstract:Context-aware decision support in the operating room can foster surgical safety and efficiency by leveraging real-time feedback from surgical workflow analysis. Most existing works recognize surgical activities at a coarse-grained level, such as phases, steps or events, leaving out fine-grained interaction details about the surgical activity; yet those are needed for more helpful AI assistance in the operating room. Recognizing surgical actions as triplets of <instrument, verb, target> combination delivers comprehensive details about the activities taking place in surgical videos. This paper presents CholecTriplet2021: an endoscopic vision challenge organized at MICCAI 2021 for the recognition of surgical action triplets in laparoscopic videos. The challenge granted private access to the large-scale CholecT50 dataset, which is annotated with action triplet information. In this paper, we present the challenge setup and assessment of the state-of-the-art deep learning methods proposed by the participants during the challenge. A total of 4 baseline methods from the challenge organizers and 19 new deep learning algorithms by competing teams are presented to recognize surgical action triplets directly from surgical videos, achieving mean average precision (mAP) ranging from 4.2% to 38.1%. This study also analyzes the significance of the results obtained by the presented approaches, performs a thorough methodological comparison between them, in-depth result analysis, and proposes a novel ensemble method for enhanced recognition. Our analysis shows that surgical workflow analysis is not yet solved, and also highlights interesting directions for future research on fine-grained surgical activity recognition which is of utmost importance for the development of AI in surgery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge