Nataša Petrović
Enhancing Generalization in Sickle Cell Disease Diagnosis through Ensemble Methods and Feature Importance Analysis
Jan 19, 2026Abstract:This work presents a novel approach for selecting the optimal ensemble-based classification method and features with a primarly focus on achieving generalization, based on the state-of-the-art, to provide diagnostic support for Sickle Cell Disease using peripheral blood smear images of red blood cells. We pre-processed and segmented the microscopic images to ensure the extraction of high-quality features. To ensure the reliability of our proposed system, we conducted an in-depth analysis of interpretability. Leveraging techniques established in the literature, we extracted features from blood cells and employed ensemble machine learning methods to classify their morphology. Furthermore, we have devised a methodology to identify the most critical features for classification, aimed at reducing complexity and training time and enhancing interpretability in opaque models. Lastly, we validated our results using a new dataset, where our model overperformed state-of-the-art models in terms of generalization. The results of classifier ensembled of Random Forest and Extra Trees classifier achieved an harmonic mean of precision and recall (F1-score) of 90.71\% and a Sickle Cell Disease diagnosis support score (SDS-score) of 93.33\%. These results demonstrate notable enhancement from previous ones with Gradient Boosting classifier (F1-score 87.32\% and SDS-score 89.51\%). To foster scientific progress, we have made available the parameters for each model, the implemented code library, and the confusion matrices with the raw data.
Crowdsourced human-based computational approach for tagging peripheral blood smear sample images from Sickle Cell Disease patients using non-expert users
Jan 13, 2025Abstract:In this paper, we present a human-based computation approach for the analysis of peripheral blood smear (PBS) images images in patients with Sickle Cell Disease (SCD). We used the Mechanical Turk microtask market to crowdsource the labeling of PBS images. We then use the expert-tagged erythrocytesIDB dataset to assess the accuracy and reliability of our proposal. Our results showed that when a robust consensus is achieved among the Mechanical Turk workers, probability of error is very low, based on comparison with expert analysis. This suggests that our proposed approach can be used to annotate datasets of PBS images, which can then be used to train automated methods for the diagnosis of SCD. In future work, we plan to explore the potential integration of our findings with outcomes obtained through automated methodologies. This could lead to the development of more accurate and reliable methods for the diagnosis of SCD
Sickle-cell disease diagnosis support selecting the most appropriate machinelearning method: Towards a general and interpretable approach for cellmorphology analysis from microscopy images
Oct 09, 2020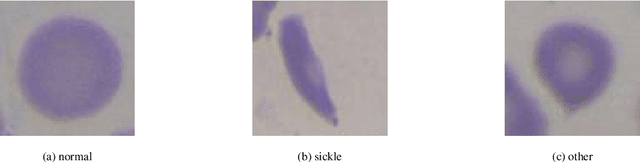

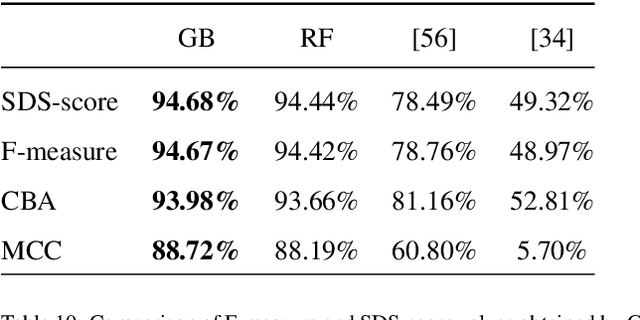
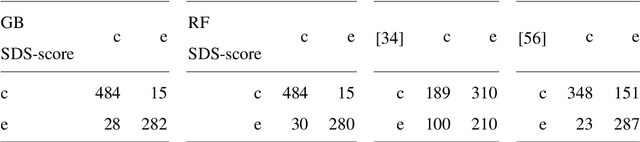
Abstract:In this work we propose an approach to select the classification method and features, based on the state-of-the-art, with best performance for diagnostic support through peripheral blood smear images of red blood cells. In our case we used samples of patients with sickle-cell disease which can be generalized for other study cases. To trust the behavior of the proposed system, we also analyzed the interpretability. We pre-processed and segmented microscopic images, to ensure high feature quality. We applied the methods used in the literature to extract the features from blood cells and the machine learning methods to classify their morphology. Next, we searched for their best parameters from the resulting data in the feature extraction phase. Then, we found the best parameters for every classifier using Randomized and Grid search. For the sake of scientific progress, we published parameters for each classifier, the implemented code library, the confusion matrices with the raw data, and we used the public erythrocytesIDB dataset for validation. We also defined how to select the most important features for classification to decrease the complexity and the training time, and for interpretability purpose in opaque models. Finally, comparing the best performing classification methods with the state-of-the-art, we obtained better results even with interpretable model classifiers.
* 35 pages, 10 tables
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge