Manuel González-Hidalgo
Diagnosis Support of Sickle Cell Anemia by Classifying Red Blood Cell Shape in Peripheral Blood Images
Jan 19, 2026Abstract:Red blood cell (RBC) deformation is the consequence of several diseases, including sickle cell anemia, which causes recurring episodes of pain and severe pronounced anemia. Monitoring patients with these diseases involves the observation of peripheral blood samples under a microscope, a time-consuming procedure. Moreover, a specialist is required to perform this technique, and owing to the subjective nature of the observation of isolated RBCs, the error rate is high. In this paper, we propose an automated method for differentially enumerating RBCs that uses peripheral blood smear image analysis. In this method, the objects of interest in the image are segmented using a Chan-Vese active contour model. An analysis is then performed to classify the RBCs, also called erythrocytes, as normal or elongated or having other deformations, using the basic shape analysis descriptors: circular shape factor (CSF) and elliptical shape factor (ESF). To analyze cells that become partially occluded in a cluster during sample preparation, an elliptical adjustment is performed to allow the analysis of erythrocytes with discoidal and elongated shapes. The images of patient blood samples used in the study were acquired by a clinical laboratory specialist in the Special Hematology Department of the ``Dr. Juan Bruno Zayas'' General Hospital in Santiago de Cuba. A comparison of the results obtained by the proposed method in our experiments with those obtained by some state-of-the-art methods showed that the proposed method is superior for the diagnosis of sickle cell anemia. This superiority is achieved for evidenced by the obtained F-measure value (0.97 for normal cells and 0.95 for elongated ones) and several overall multiclass performance measures. The results achieved by the proposed method are suitable for the purpose of clinical treatment and diagnostic support of sickle cell anemia.
An efficient heuristic for geometric analysis of cell deformations
Jan 19, 2026Abstract:Sickle cell disease causes erythrocytes to become sickle-shaped, affecting their movement in the bloodstream and reducing oxygen delivery. It has a high global prevalence and places a significant burden on healthcare systems, especially in resource-limited regions. Automated classification of sickle cells in blood images is crucial, allowing the specialist to reduce the effort required and avoid errors when quantifying the deformed cells and assessing the severity of a crisis. Recent studies have proposed various erythrocyte representation and classification methods. Since classification depends solely on cell shape, a suitable approach models erythrocytes as closed planar curves in shape space. This approach employs elastic distances between shapes, which are invariant under rotations, translations, scaling, and reparameterizations, ensuring consistent distance measurements regardless of the curves' position, starting point, or traversal speed. While previous methods exploiting shape space distances had achieved high accuracy, we refined the model by considering the geometric characteristics of healthy and sickled erythrocytes. Our method proposes (1) to employ a fixed parameterization based on the major axis of each cell to compute distances and (2) to align each cell with two templates using this parameterization before computing distances. Aligning shapes to templates before distance computation, a concept successfully applied in areas such as molecular dynamics, and using a fixed parameterization, instead of minimizing distances across all possible parameterizations, simplifies calculations. This strategy achieves 96.03\% accuracy rate in both supervised classification and unsupervised clustering. Our method ensures efficient erythrocyte classification, maintaining or improving accuracy over shape space models while significantly reducing computational costs.
An Encoder-Decoder CNN for Hair Removal in Dermoscopic Images
Oct 10, 2020
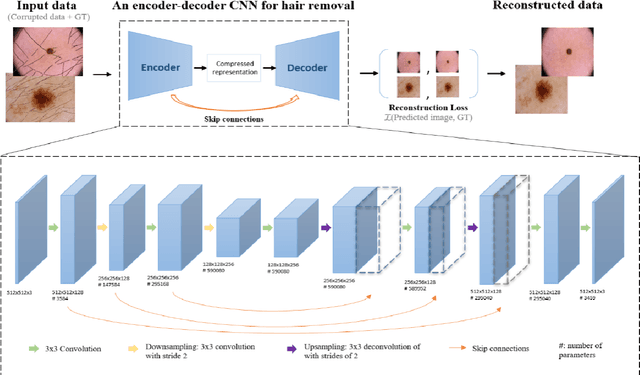
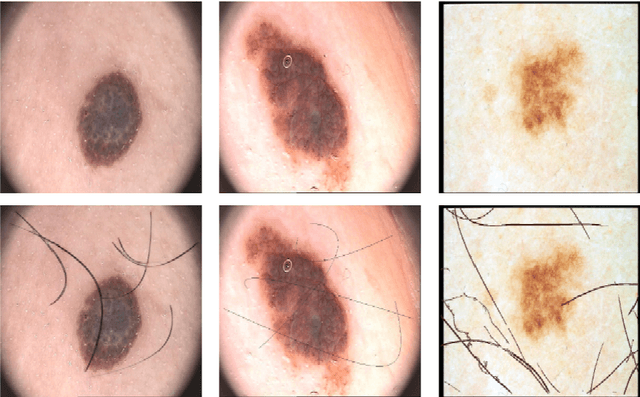
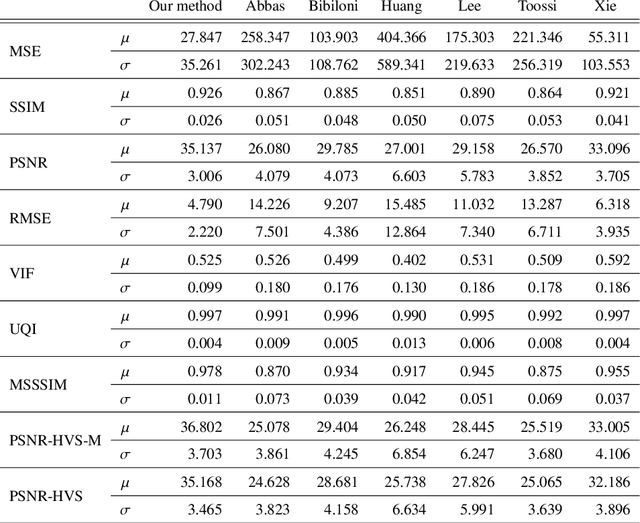
Abstract:The process of removing occluding hair has a relevant role in the early and accurate diagnosis of skin cancer. It consists of detecting hairs and restore the texture below them, which is sporadically occluded. In this work, we present a model based on convolutional neural networks for hair removal in dermoscopic images. During the network's training, we use a combined loss function to improve the restoration ability of the proposed model. In order to train the CNN and to quantitatively validate their performance, we simulate the presence of skin hair in hairless images extracted from publicly known datasets such as the PH2, dermquest, dermis, EDRA2002, and the ISIC Data Archive. As far as we know, there is no other hair removal method based on deep learning. Thus, we compare our results with six state-of-the-art algorithms based on traditional computer vision techniques by means of similarity measures that compare the reference hairless image and the one with hair simulated. Finally, a statistical test is used to compare the methods. Both qualitative and quantitative results demonstrate the effectiveness of our network.
Sickle-cell disease diagnosis support selecting the most appropriate machinelearning method: Towards a general and interpretable approach for cellmorphology analysis from microscopy images
Oct 09, 2020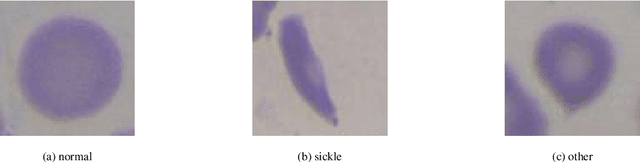

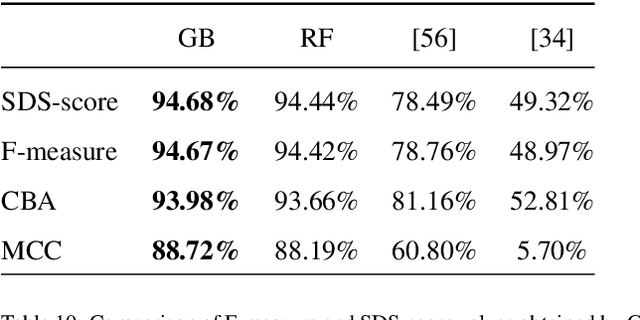
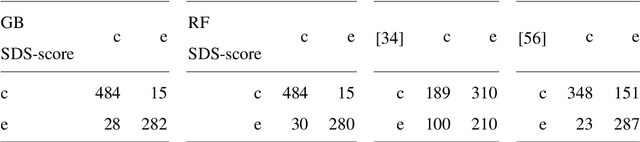
Abstract:In this work we propose an approach to select the classification method and features, based on the state-of-the-art, with best performance for diagnostic support through peripheral blood smear images of red blood cells. In our case we used samples of patients with sickle-cell disease which can be generalized for other study cases. To trust the behavior of the proposed system, we also analyzed the interpretability. We pre-processed and segmented microscopic images, to ensure high feature quality. We applied the methods used in the literature to extract the features from blood cells and the machine learning methods to classify their morphology. Next, we searched for their best parameters from the resulting data in the feature extraction phase. Then, we found the best parameters for every classifier using Randomized and Grid search. For the sake of scientific progress, we published parameters for each classifier, the implemented code library, the confusion matrices with the raw data, and we used the public erythrocytesIDB dataset for validation. We also defined how to select the most important features for classification to decrease the complexity and the training time, and for interpretability purpose in opaque models. Finally, comparing the best performing classification methods with the state-of-the-art, we obtained better results even with interpretable model classifiers.
* 35 pages, 10 tables
Segmenting overlapped objects in images. A study to support the diagnosis of sickle cell disease
Aug 03, 2020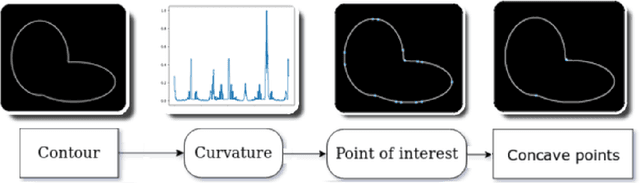
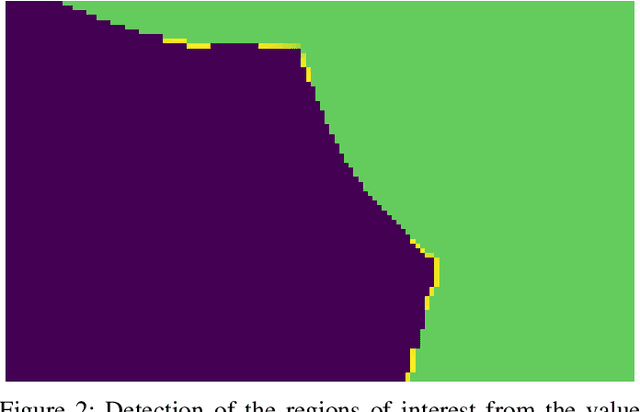
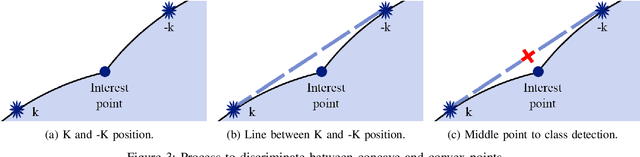
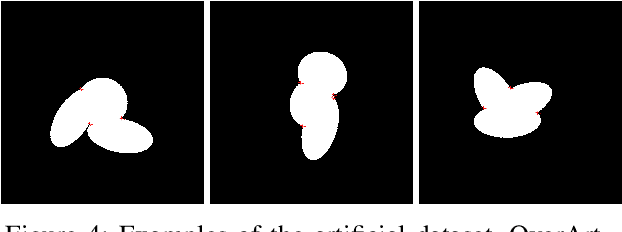
Abstract:Overlapped objects are found on multiple kinds of images, they are a source of problem due its partial information. Multiple types of algorithm are used to address this problem from simple and naive methods to more complex ones. In this work we propose a new method for the segmentation of overlapped object. Finally we compare the results of this algorithm with the state-of-art in two experiments: one with a new dataset, developed specially for this work, and red blood smears from sickle-cell disease patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge