Narges Razavian
BERT-XML: Large Scale Automated ICD Coding Using BERT Pretraining
May 26, 2020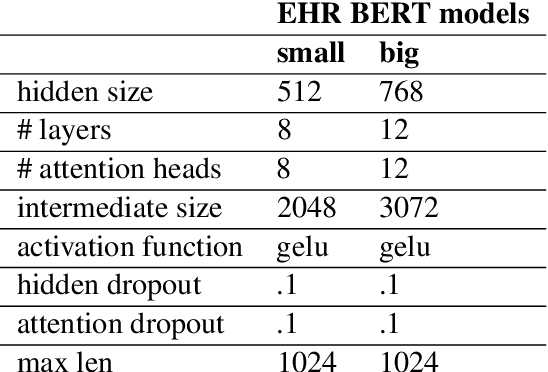
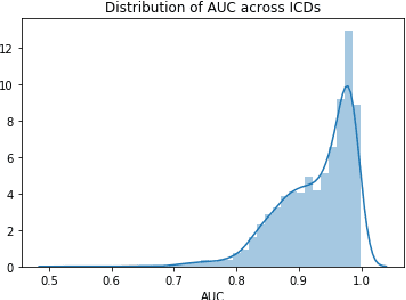
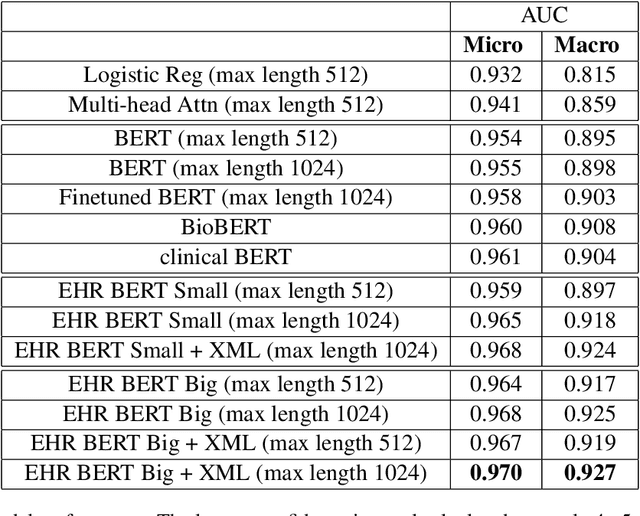
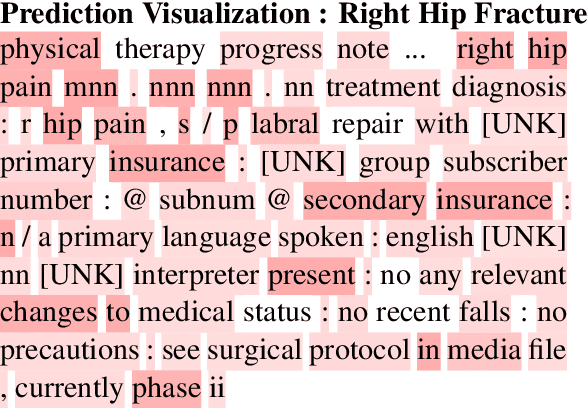
Abstract:Clinical interactions are initially recorded and documented in free text medical notes. ICD coding is the task of classifying and coding all diagnoses, symptoms and procedures associated with a patient's visit. The process is often manual and extremely time-consuming and expensive for hospitals. In this paper, we propose a machine learning model, BERT-XML, for large scale automated ICD coding from EHR notes, utilizing recently developed unsupervised pretraining that have achieved state of the art performance on a variety of NLP tasks. We train a BERT model from scratch on EHR notes, learning with vocabulary better suited for EHR tasks and thus outperform off-the-shelf models. We adapt the BERT architecture for ICD coding with multi-label attention. While other works focus on small public medical datasets, we have produced the first large scale ICD-10 classification model using millions of EHR notes to predict thousands of unique ICD codes.
Graph Neural Network on Electronic Health Records for Predicting Alzheimer's Disease
Dec 08, 2019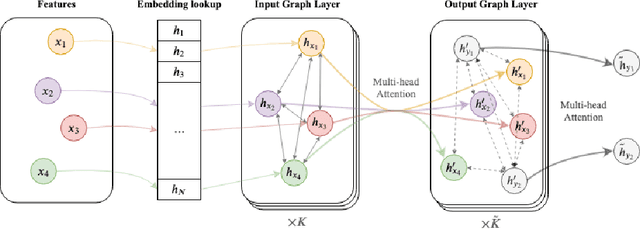
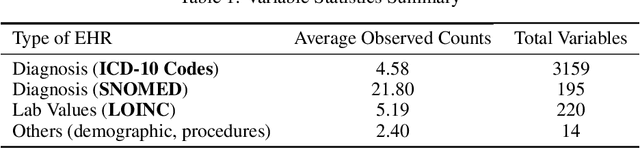
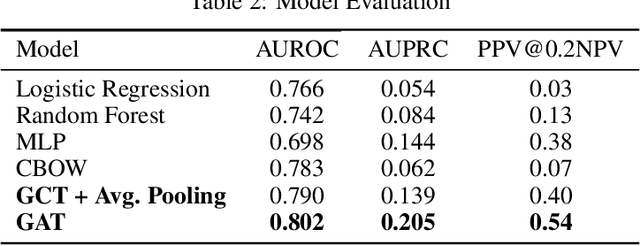
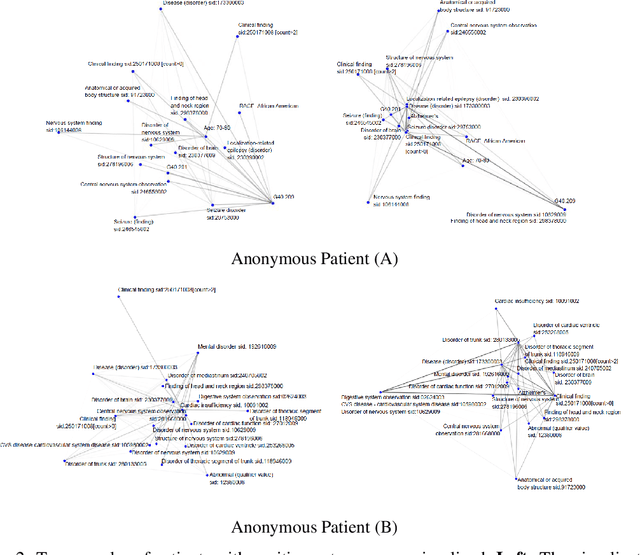
Abstract:The cause of Alzheimer's disease (AD) is poorly understood, so forecasting AD remains a hard task in population health. Failure of clinical trials for AD treatments indicates that AD should be intervened at the earlier, pre-symptomatic stages. Developing an explainable method for predicting AD is critical for providing better treatment targets, better clinical trial recruitment, and better clinical care for the AD patients. In this paper, we present a novel approach for disease (AD) prediction based on Electronic Health Records (EHR) and graph neural network. Our method improves the performance on sparse data which is common in EHR, and obtains state-of-art results in predicting AD 12 to 24 months in advance on real-world EHR data, compared to other baseline results. Our approach also provides an insight into the structural relationship among different diagnosis, Lab values, and procedures from EHR as per graph structures learned by our model.
Tracing State-Level Obesity Prevalence from Sentence Embeddings of Tweets: A Feasibility Study
Dec 02, 2019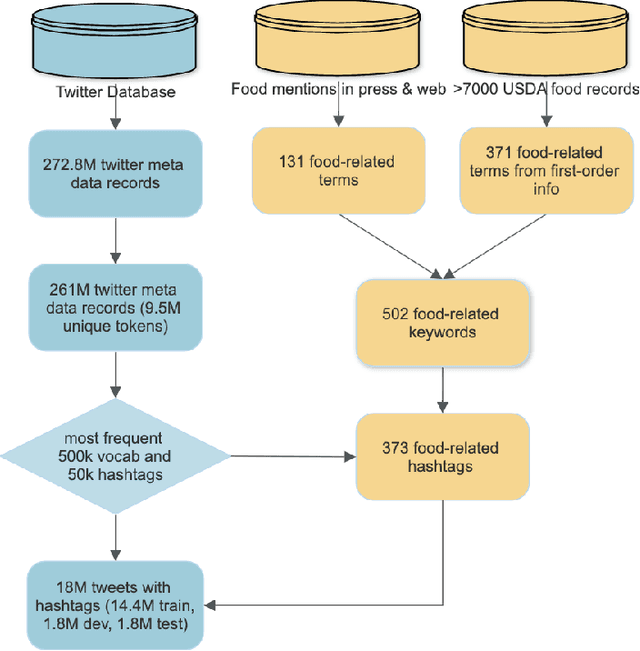

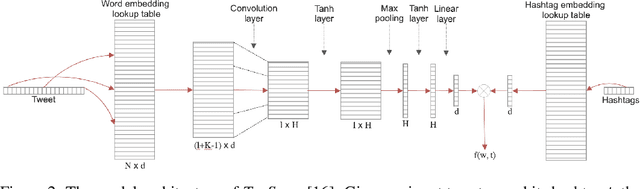
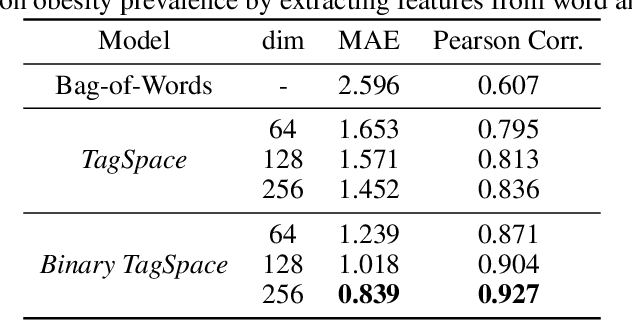
Abstract:Twitter data has been shown broadly applicable for public health surveillance. Previous public health studies based on Twitter data have largely relied on keyword-matching or topic models for clustering relevant tweets. However, both methods suffer from the short-length of texts and unpredictable noise that naturally occurs in user-generated contexts. In response, we introduce a deep learning approach that uses hashtags as a form of supervision and learns tweet embeddings for extracting informative textual features. In this case study, we address the specific task of estimating state-level obesity from dietary-related textual features. Our approach yields an estimation that strongly correlates the textual features to government data and outperforms the keyword-matching baseline. The results also demonstrate the potential of discovering risk factors using the textual features. This method is general-purpose and can be applied to a wide range of Twitter-based public health studies.
Towards Quantification of Bias in Machine Learning for Healthcare: A Case Study of Renal Failure Prediction
Nov 18, 2019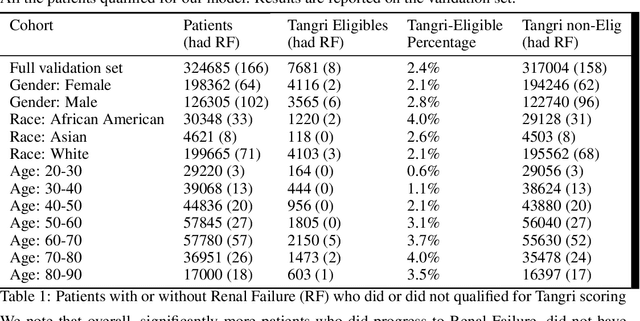
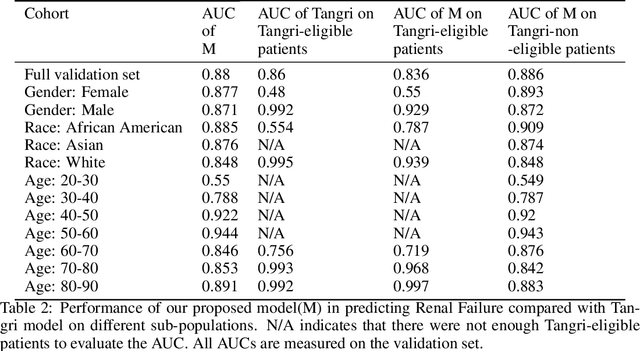
Abstract:As machine learning (ML) models, trained on real-world datasets, become common practice, it is critical to measure and quantify their potential biases. In this paper, we focus on renal failure and compare a commonly used traditional risk score, Tangri, with a more powerful machine learning model, which has access to a larger variable set and trained on 1.6 million patients' EHR data. We will compare and discuss the generalization and applicability of these two models, in an attempt to quantify biases of status quo clinical practice, compared to ML-driven models.
DARTS: DenseUnet-based Automatic Rapid Tool for brain Segmentation
Nov 14, 2019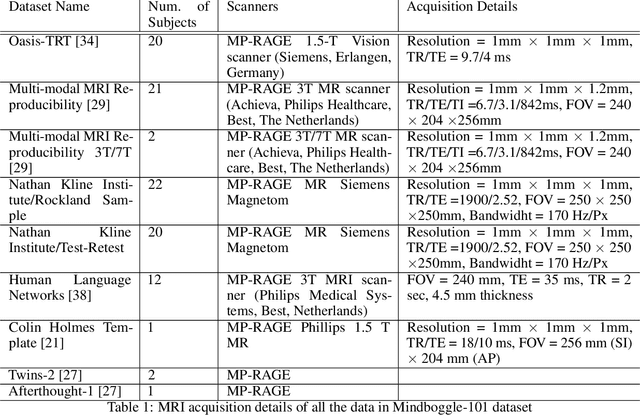
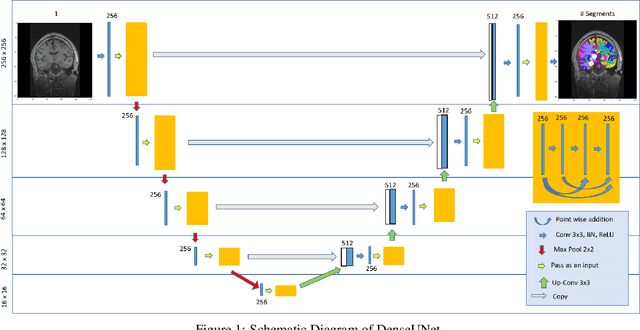
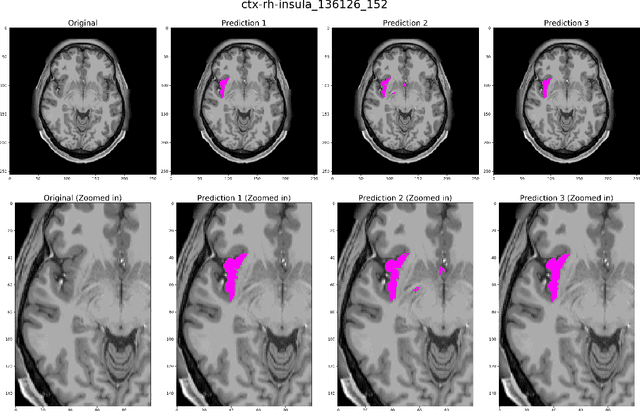
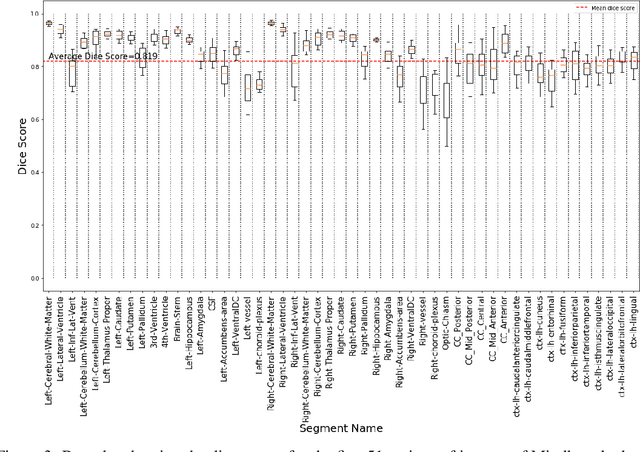
Abstract:Quantitative, volumetric analysis of Magnetic Resonance Imaging (MRI) is a fundamental way researchers study the brain in a host of neurological conditions including normal maturation and aging. Despite the availability of open-source brain segmentation software, widespread clinical adoption of volumetric analysis has been hindered due to processing times and reliance on manual corrections. Here, we extend the use of deep learning models from proof-of-concept, as previously reported, to present a comprehensive segmentation of cortical and deep gray matter brain structures matching the standard regions of aseg+aparc included in the commonly used open-source tool, Freesurfer. The work presented here provides a real-life, rapid deep learning-based brain segmentation tool to enable clinical translation as well as research application of quantitative brain segmentation. The advantages of the presented tool include short (~1 minute) processing time and improved segmentation quality. This is the first study to perform quick and accurate segmentation of 102 brain regions based on the surface-based protocol (DMK protocol), widely used by experts in the field. This is also the first work to include an expert reader study to assess the quality of the segmentation obtained using a deep-learning-based model. We show the superior performance of our deep-learning-based models over the traditional segmentation tool, Freesurfer. We refer to the proposed deep learning-based tool as DARTS (DenseUnet-based Automatic Rapid Tool for brain Segmentation). Our tool and trained models are available at https://github.com/NYUMedML/DARTS
On the design of convolutional neural networks for automatic detection of Alzheimer's disease
Nov 12, 2019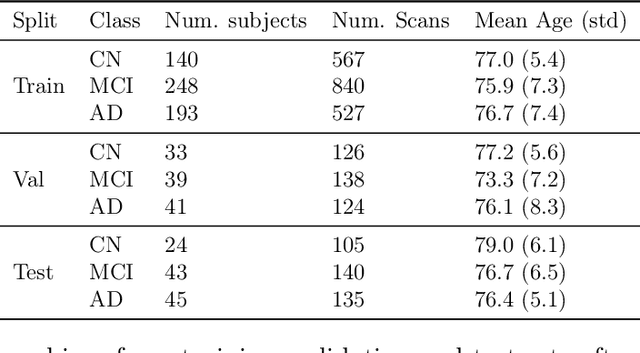
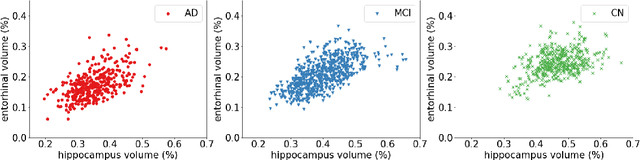
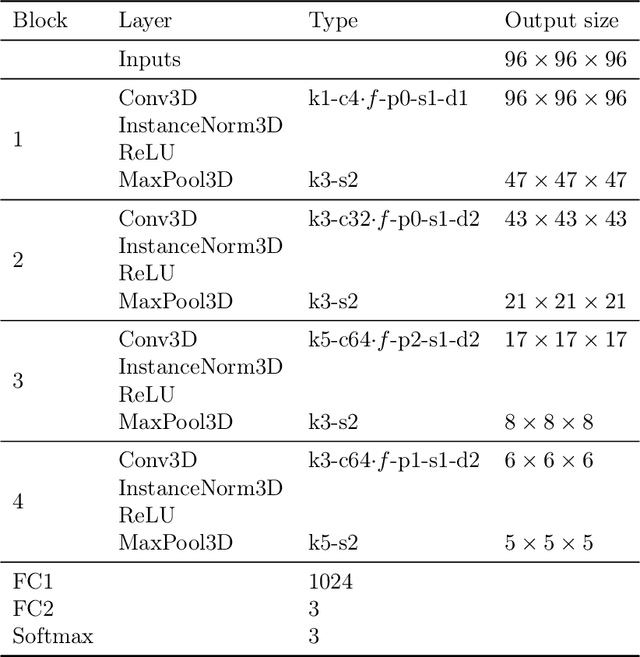
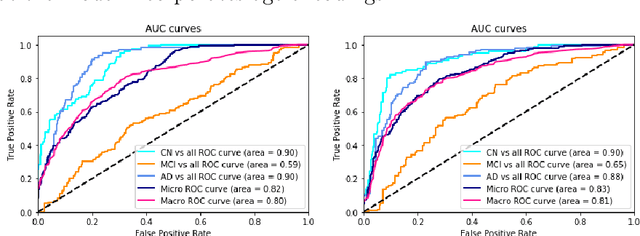
Abstract:Early detection is a crucial goal in the study of Alzheimer's Disease (AD). In this work, we describe several techniques to boost the performance of 3D convolutional neural networks trained to detect AD using structural brain MRI scans. Specifically, we provide evidence that (1) instance normalization outperforms batch normalization, (2) early spatial downsampling negatively affects performance, (3) widening the model brings consistent gains while increasing the depth does not, and (4) incorporating age information yields moderate improvement. Together, these insights yield an increment of approximately 14% in test accuracy over existing models when distinguishing between patients with AD, mild cognitive impairment, and controls in the ADNI dataset. Similar performance is achieved on an independent dataset.
* Machine Learning for Health Workshop, NeurIPS2019. Authors Fernandez-Granda and Razavian are joint last authors
Deep EHR: Chronic Disease Prediction Using Medical Notes
Aug 15, 2018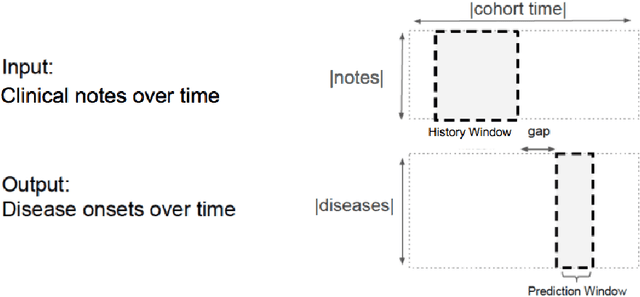

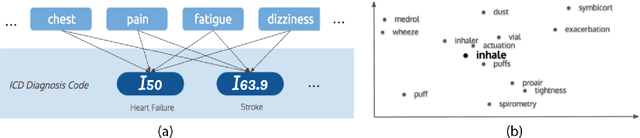
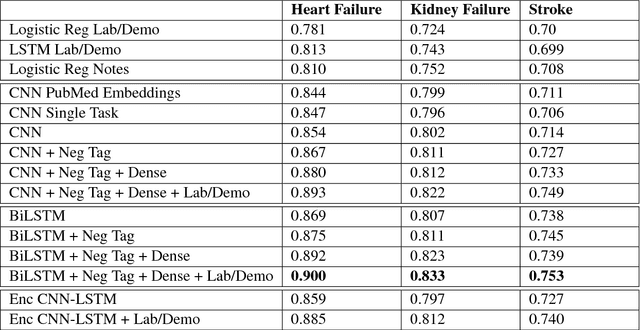
Abstract:Early detection of preventable diseases is important for better disease management, improved inter-ventions, and more efficient health-care resource allocation. Various machine learning approacheshave been developed to utilize information in Electronic Health Record (EHR) for this task. Majorityof previous attempts, however, focus on structured fields and lose the vast amount of information inthe unstructured notes. In this work we propose a general multi-task framework for disease onsetprediction that combines both free-text medical notes and structured information. We compareperformance of different deep learning architectures including CNN, LSTM and hierarchical models.In contrast to traditional text-based prediction models, our approach does not require disease specificfeature engineering, and can handle negations and numerical values that exist in the text. Ourresults on a cohort of about 1 million patients show that models using text outperform modelsusing just structured data, and that models capable of using numerical values and negations in thetext, in addition to the raw text, further improve performance. Additionally, we compare differentvisualization methods for medical professionals to interpret model predictions.
Multi-task Prediction of Disease Onsets from Longitudinal Lab Tests
Sep 20, 2016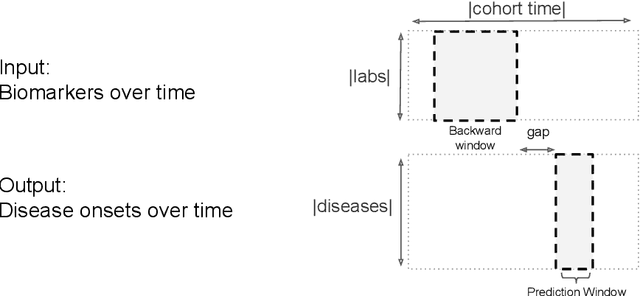
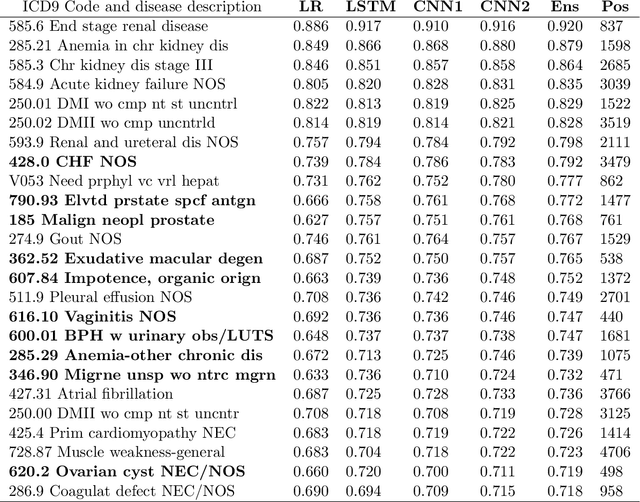
Abstract:Disparate areas of machine learning have benefited from models that can take raw data with little preprocessing as input and learn rich representations of that raw data in order to perform well on a given prediction task. We evaluate this approach in healthcare by using longitudinal measurements of lab tests, one of the more raw signals of a patient's health state widely available in clinical data, to predict disease onsets. In particular, we train a Long Short-Term Memory (LSTM) recurrent neural network and two novel convolutional neural networks for multi-task prediction of disease onset for 133 conditions based on 18 common lab tests measured over time in a cohort of 298K patients derived from 8 years of administrative claims data. We compare the neural networks to a logistic regression with several hand-engineered, clinically relevant features. We find that the representation-based learning approaches significantly outperform this baseline. We believe that our work suggests a new avenue for patient risk stratification based solely on lab results.
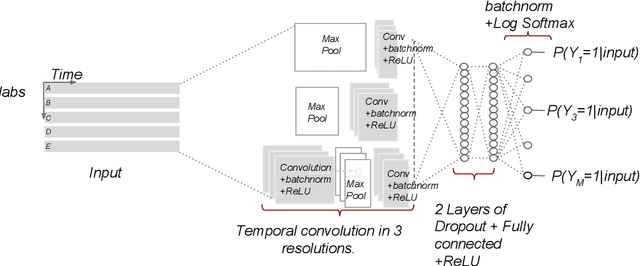
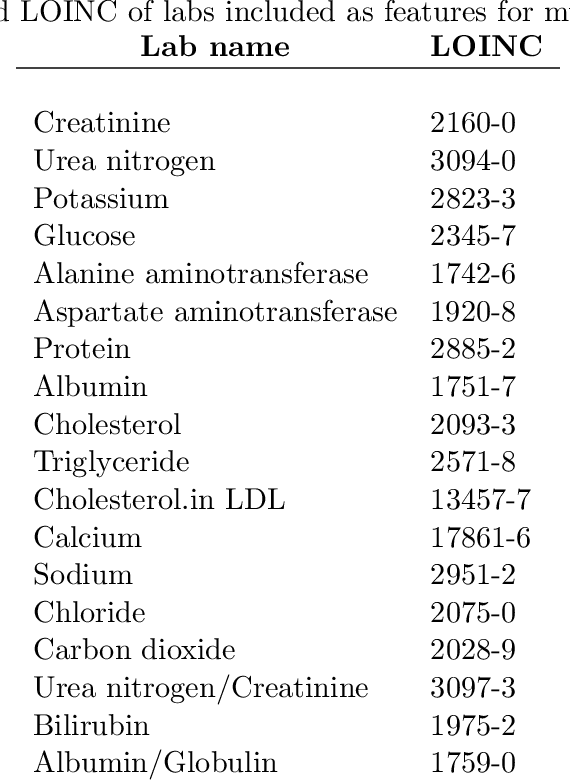
Temporal Convolutional Neural Networks for Diagnosis from Lab Tests
Mar 11, 2016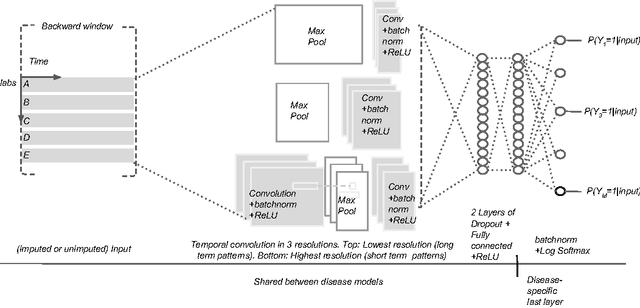
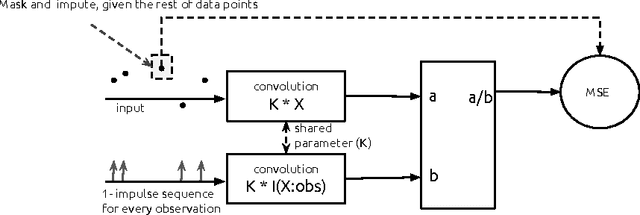
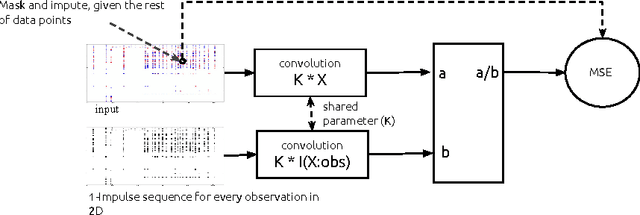
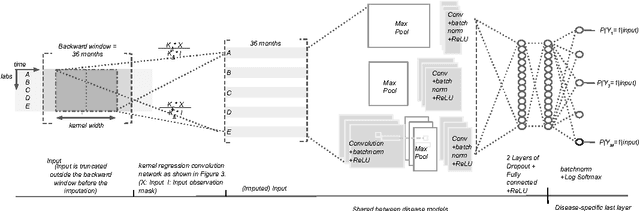
Abstract:Early diagnosis of treatable diseases is essential for improving healthcare, and many diseases' onsets are predictable from annual lab tests and their temporal trends. We introduce a multi-resolution convolutional neural network for early detection of multiple diseases from irregularly measured sparse lab values. Our novel architecture takes as input both an imputed version of the data and a binary observation matrix. For imputing the temporal sparse observations, we develop a flexible, fast to train method for differentiable multivariate kernel regression. Our experiments on data from 298K individuals over 8 years, 18 common lab measurements, and 171 diseases show that the temporal signatures learned via convolution are significantly more predictive than baselines commonly used for early disease diagnosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge