Matthias W. Beckmann
Unsupervised Anomaly Detection of Diseases in the Female Pelvis for Real-Time MR Imaging
Feb 05, 2026Abstract:Pelvic diseases in women of reproductive age represent a major global health burden, with diagnosis frequently delayed due to high anatomical variability, complicating MRI interpretation. Existing AI approaches are largely disease-specific and lack real-time compatibility, limiting generalizability and clinical integration. To address these challenges, we establish a benchmark framework for disease- and parameter-agnostic, real-time-compatible unsupervised anomaly detection in pelvic MRI. The method uses a residual variational autoencoder trained exclusively on healthy sagittal T2-weighted scans acquired across diverse imaging protocols to model normal pelvic anatomy. During inference, reconstruction error heatmaps indicate deviations from learned healthy structure, enabling detection of pathological regions without labeled abnormal data. The model is trained on 294 healthy scans and augmented with diffusion-generated synthetic data to improve robustness. Quantitative evaluation on the publicly available Uterine Myoma MRI Dataset yields an average area-under-the-curve (AUC) value of 0.736, with 0.828 sensitivity and 0.692 specificity. Additional inter-observer clinical evaluation extends analysis to endometrial cancer, endometriosis, and adenomyosis, revealing the influence of anatomical heterogeneity and inter-observer variability on performance interpretation. With a reconstruction time of approximately 92.6 frames per second, the proposed framework establishes a baseline for unsupervised anomaly detection in the female pelvis and supports future integration into real-time MRI. Code is available upon request (https://github.com/AniKnu/UADPelvis), prospective data sets are available for academic collaboration.
Improved HER2 Tumor Segmentation with Subtype Balancing using Deep Generative Networks
Nov 11, 2022Abstract:Tumor segmentation in histopathology images is often complicated by its composition of different histological subtypes and class imbalance. Oversampling subtypes with low prevalence features is not a satisfactory solution since it eventually leads to overfitting. We propose to create synthetic images with semantically-conditioned deep generative networks and to combine subtype-balanced synthetic images with the original dataset to achieve better segmentation performance. We show the suitability of Generative Adversarial Networks (GANs) and especially diffusion models to create realistic images based on subtype-conditioning for the use case of HER2-stained histopathology. Additionally, we show the capability of diffusion models to conditionally inpaint HER2 tumor areas with modified subtypes. Combining the original dataset with the same amount of diffusion-generated images increased the tumor Dice score from 0.833 to 0.854 and almost halved the variance between the HER2 subtype recalls. These results create the basis for more reliable automatic HER2 analysis with lower performance variance between individual HER2 subtypes.
Superpixel Pre-Segmentation of HER2 Slides for Efficient Annotation
Jan 19, 2022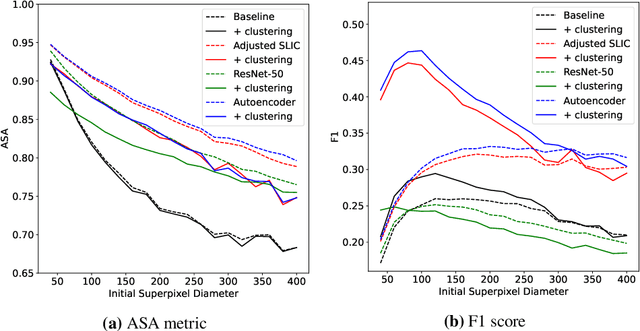
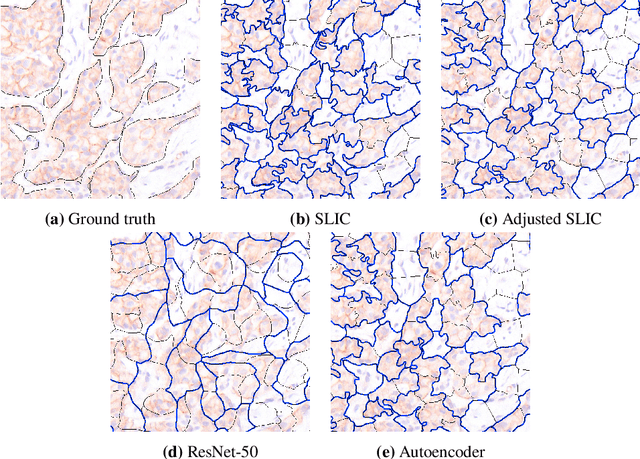
Abstract:Supervised deep learning has shown state-of-the-art performance for medical image segmentation across different applications, including histopathology and cancer research; however, the manual annotation of such data is extremely laborious. In this work, we explore the use of superpixel approaches to compute a pre-segmentation of HER2 stained images for breast cancer diagnosis that facilitates faster manual annotation and correction in a second step. Four methods are compared: Standard Simple Linear Iterative Clustering (SLIC) as a baseline, a domain adapted SLIC, and superpixels based on feature embeddings of a pretrained ResNet-50 and a denoising autoencoder. To tackle oversegmentation, we propose to hierarchically merge superpixels, based on their content in the respective feature space. When evaluating the approaches on fully manually annotated images, we observe that the autoencoder-based superpixels achieve a 23% increase in boundary F1 score compared to the baseline SLIC superpixels. Furthermore, the boundary F1 score increases by 73% when hierarchical clustering is applied on the adapted SLIC and the autoencoder-based superpixels. These evaluations show encouraging first results for a pre-segmentation for efficient manual refinement without the need for an initial set of annotated training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge