Matthew P Lungren
Evaluation of GPT-3.5 and GPT-4 for supporting real-world information needs in healthcare delivery
May 01, 2023


Abstract:Despite growing interest in using large language models (LLMs) in healthcare, current explorations do not assess the real-world utility and safety of LLMs in clinical settings. Our objective was to determine whether two LLMs can serve information needs submitted by physicians as questions to an informatics consultation service in a safe and concordant manner. Sixty six questions from an informatics consult service were submitted to GPT-3.5 and GPT-4 via simple prompts. 12 physicians assessed the LLM responses' possibility of patient harm and concordance with existing reports from an informatics consultation service. Physician assessments were summarized based on majority vote. For no questions did a majority of physicians deem either LLM response as harmful. For GPT-3.5, responses to 8 questions were concordant with the informatics consult report, 20 discordant, and 9 were unable to be assessed. There were 29 responses with no majority on "Agree", "Disagree", and "Unable to assess". For GPT-4, responses to 13 questions were concordant, 15 discordant, and 3 were unable to be assessed. There were 35 responses with no majority. Responses from both LLMs were largely devoid of overt harm, but less than 20% of the responses agreed with an answer from an informatics consultation service, responses contained hallucinated references, and physicians were divided on what constitutes harm. These results suggest that while general purpose LLMs are able to provide safe and credible responses, they often do not meet the specific information need of a given question. A definitive evaluation of the usefulness of LLMs in healthcare settings will likely require additional research on prompt engineering, calibration, and custom-tailoring of general purpose models.
CheXstray: Real-time Multi-Modal Data Concordance for Drift Detection in Medical Imaging AI
Feb 06, 2022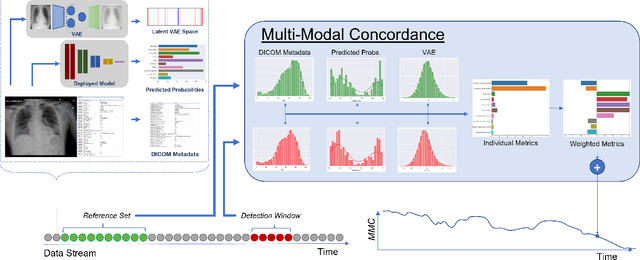
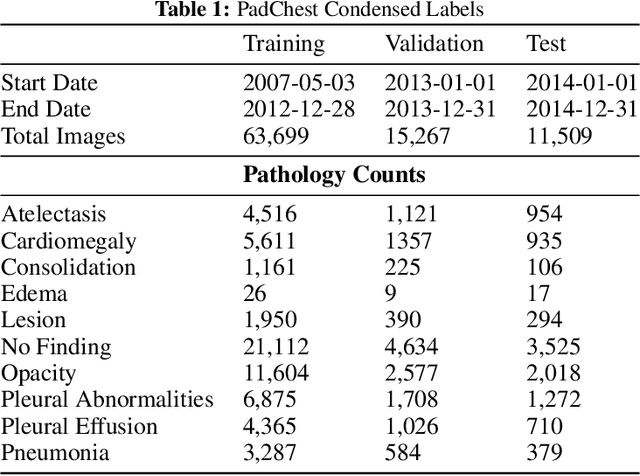
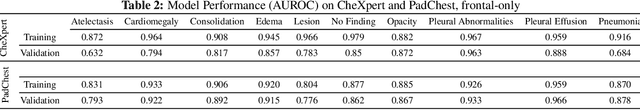
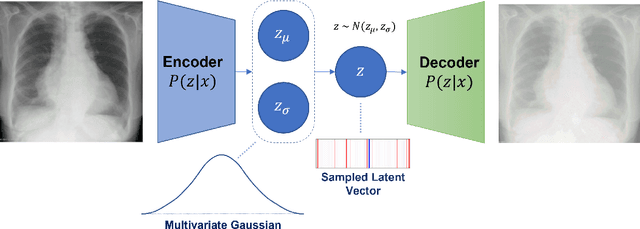
Abstract:Rapidly expanding Clinical AI applications worldwide have the potential to impact to all areas of medical practice. Medical imaging applications constitute a vast majority of approved clinical AI applications. Though healthcare systems are eager to adopt AI solutions a fundamental question remains: \textit{what happens after the AI model goes into production?} We use the CheXpert and PadChest public datasets to build and test a medical imaging AI drift monitoring workflow that tracks data and model drift without contemporaneous ground truth. We simulate drift in multiple experiments to compare model performance with our novel multi-modal drift metric, which uses DICOM metadata, image appearance representation from a variational autoencoder (VAE), and model output probabilities as input. Through experimentation, we demonstrate a strong proxy for ground truth performance using unsupervised distributional shifts in relevant metadata, predicted probabilities, and VAE latent representation. Our key contributions include (1) proof-of-concept for medical imaging drift detection including use of VAE and domain specific statistical methods (2) a multi-modal methodology for measuring and unifying drift metrics (3) new insights into the challenges and solutions for observing deployed medical imaging AI (4) creation of open-source tools enabling others to easily run their own workflows or scenarios. This work has important implications for addressing the translation gap related to continuous medical imaging AI model monitoring in dynamic healthcare environments.
TorchXRayVision: A library of chest X-ray datasets and models
Oct 31, 2021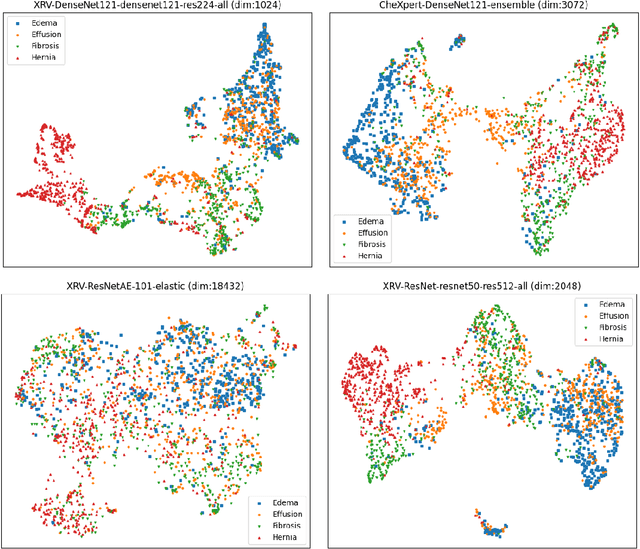
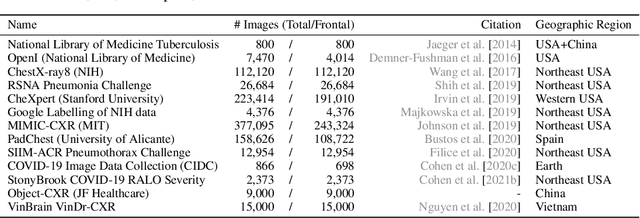
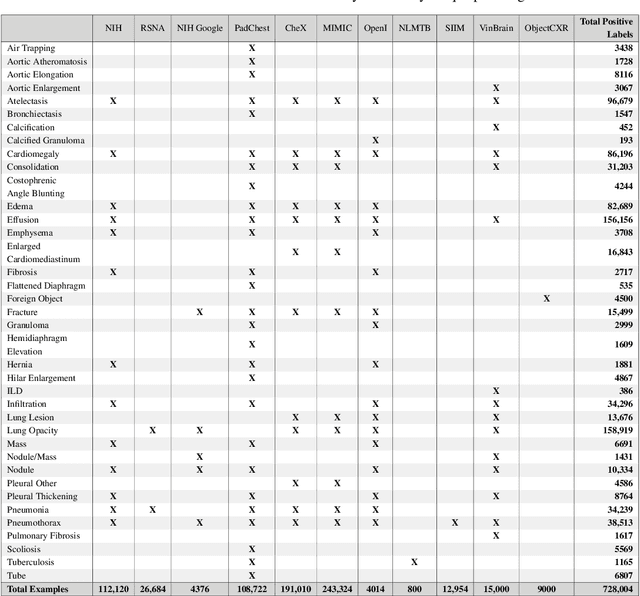
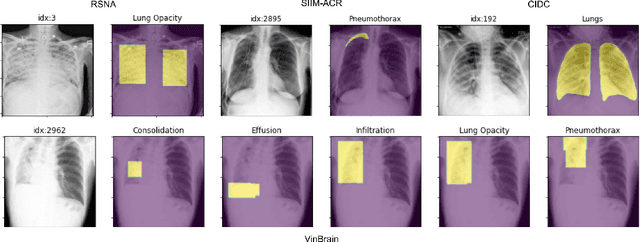
Abstract:TorchXRayVision is an open source software library for working with chest X-ray datasets and deep learning models. It provides a common interface and common pre-processing chain for a wide set of publicly available chest X-ray datasets. In addition, a number of classification and representation learning models with different architectures, trained on different data combinations, are available through the library to serve as baselines or feature extractors.
OncoNet: Weakly Supervised Siamese Network to automate cancer treatment response assessment between longitudinal FDG PET/CT examinations
Aug 03, 2021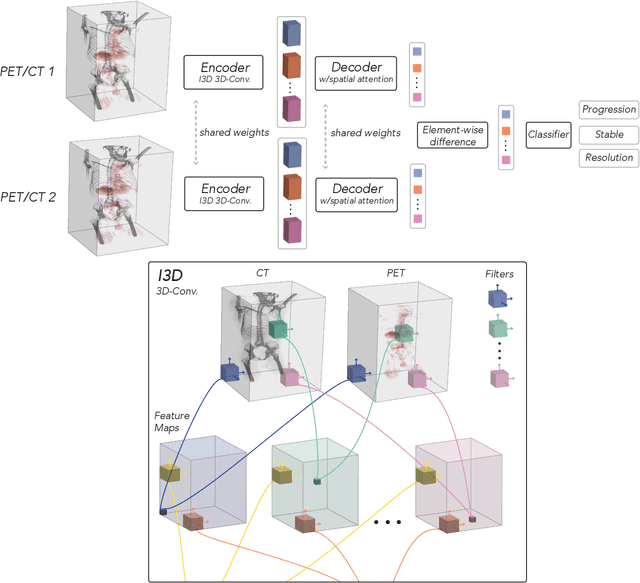

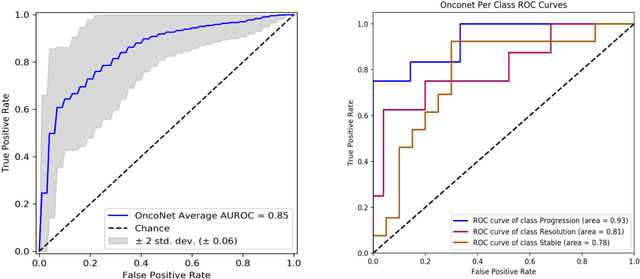
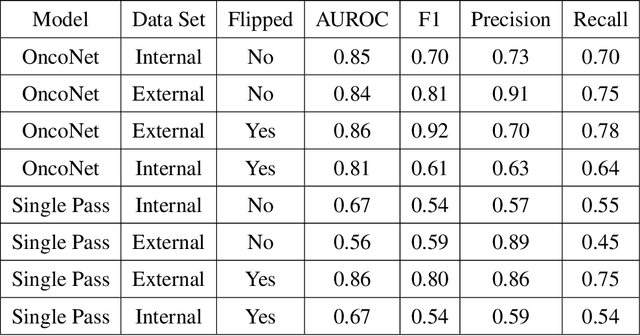
Abstract:FDG PET/CT imaging is a resource intensive examination critical for managing malignant disease and is particularly important for longitudinal assessment during therapy. Approaches to automate longtudinal analysis present many challenges including lack of available longitudinal datasets, managing complex large multimodal imaging examinations, and need for detailed annotations for traditional supervised machine learning. In this work we develop OncoNet, novel machine learning algorithm that assesses treatment response from a 1,954 pairs of sequential FDG PET/CT exams through weak supervision using the standard uptake values (SUVmax) in associated radiology reports. OncoNet demonstrates an AUROC of 0.86 and 0.84 on internal and external institution test sets respectively for determination of change between scans while also showing strong agreement to clinical scoring systems with a kappa score of 0.8. We also curated a dataset of 1,954 paired FDG PET/CT exams designed for response assessment for the broader machine learning in healthcare research community. Automated assessment of radiographic response from FDG PET/CT with OncoNet could provide clinicians with a valuable tool to rapidly and consistently interpret change over time in longitudinal multi-modal imaging exams.
Reading Race: AI Recognises Patient's Racial Identity In Medical Images
Jul 21, 2021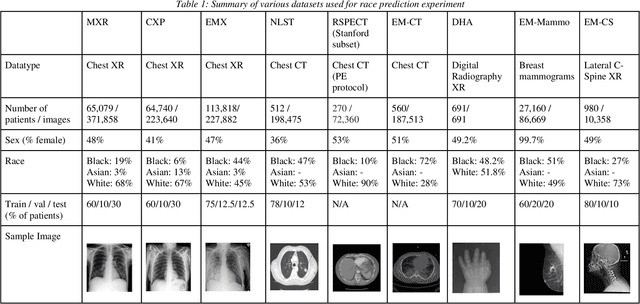
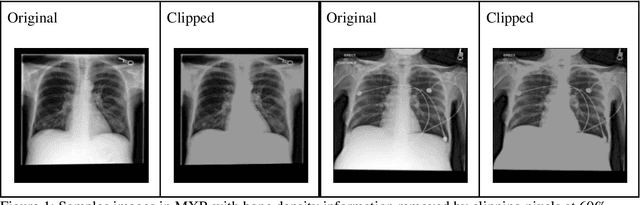
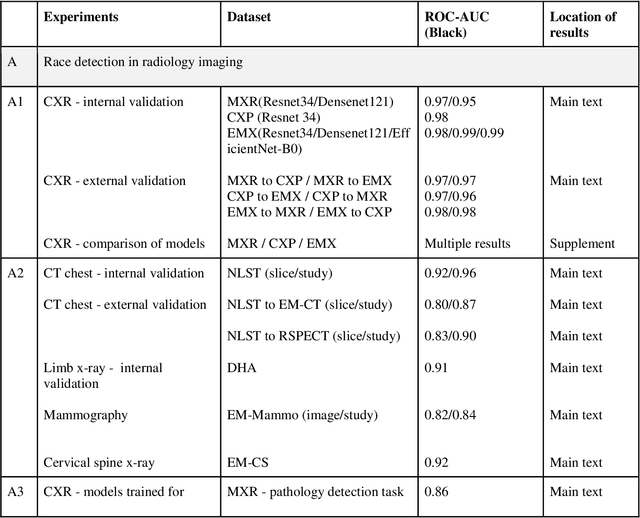
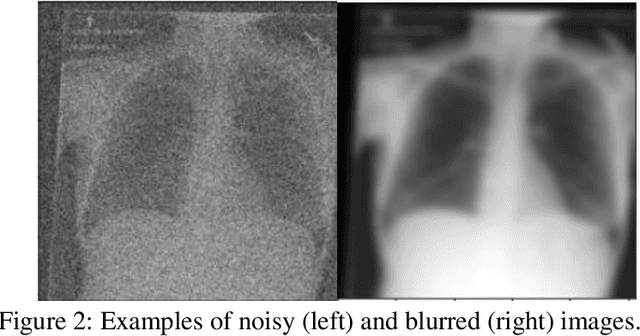
Abstract:Background: In medical imaging, prior studies have demonstrated disparate AI performance by race, yet there is no known correlation for race on medical imaging that would be obvious to the human expert interpreting the images. Methods: Using private and public datasets we evaluate: A) performance quantification of deep learning models to detect race from medical images, including the ability of these models to generalize to external environments and across multiple imaging modalities, B) assessment of possible confounding anatomic and phenotype population features, such as disease distribution and body habitus as predictors of race, and C) investigation into the underlying mechanism by which AI models can recognize race. Findings: Standard deep learning models can be trained to predict race from medical images with high performance across multiple imaging modalities. Our findings hold under external validation conditions, as well as when models are optimized to perform clinically motivated tasks. We demonstrate this detection is not due to trivial proxies or imaging-related surrogate covariates for race, such as underlying disease distribution. Finally, we show that performance persists over all anatomical regions and frequency spectrum of the images suggesting that mitigation efforts will be challenging and demand further study. Interpretation: We emphasize that model ability to predict self-reported race is itself not the issue of importance. However, our findings that AI can trivially predict self-reported race -- even from corrupted, cropped, and noised medical images -- in a setting where clinical experts cannot, creates an enormous risk for all model deployments in medical imaging: if an AI model secretly used its knowledge of self-reported race to misclassify all Black patients, radiologists would not be able to tell using the same data the model has access to.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge