Lin Fu
Multiscale Cross-Modal Mapping of Molecular, Pathologic, and Radiologic Phenotypes in Lipid-Deficient Clear Cell Renal CellCarcinoma
Dec 13, 2025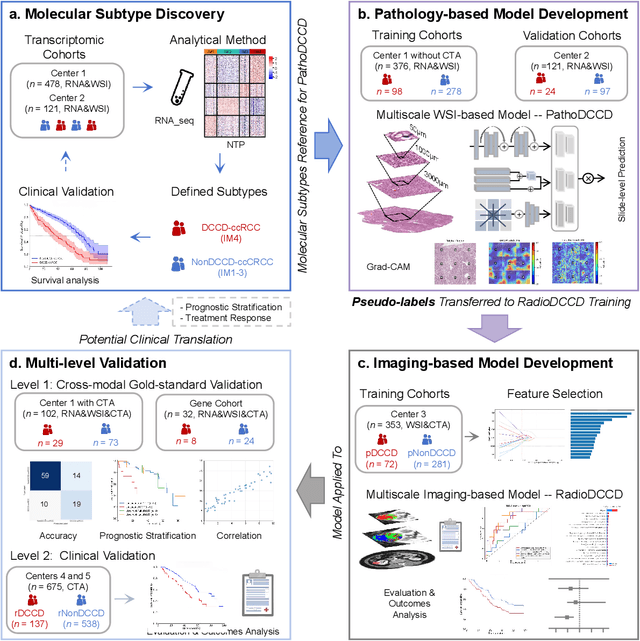
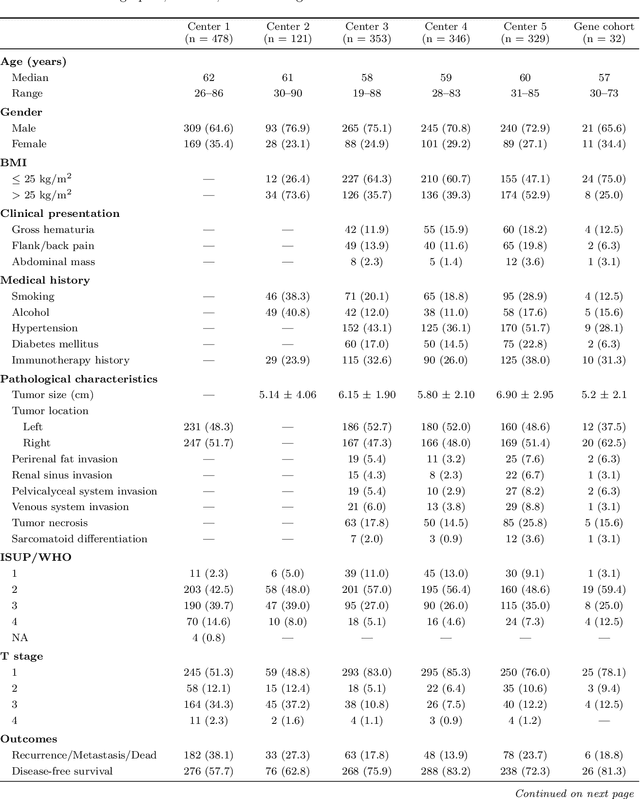
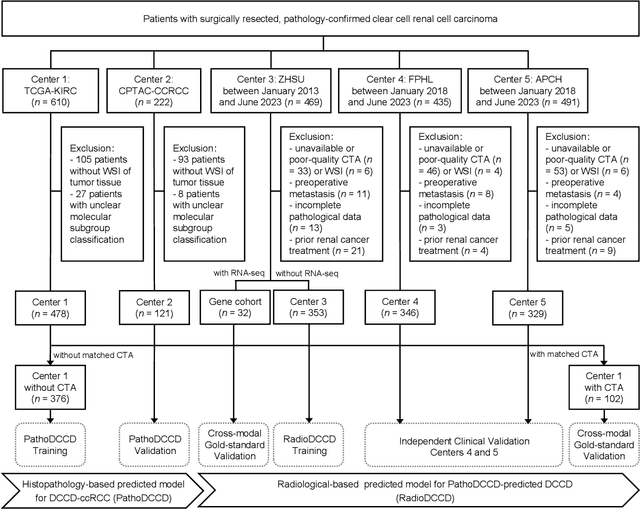
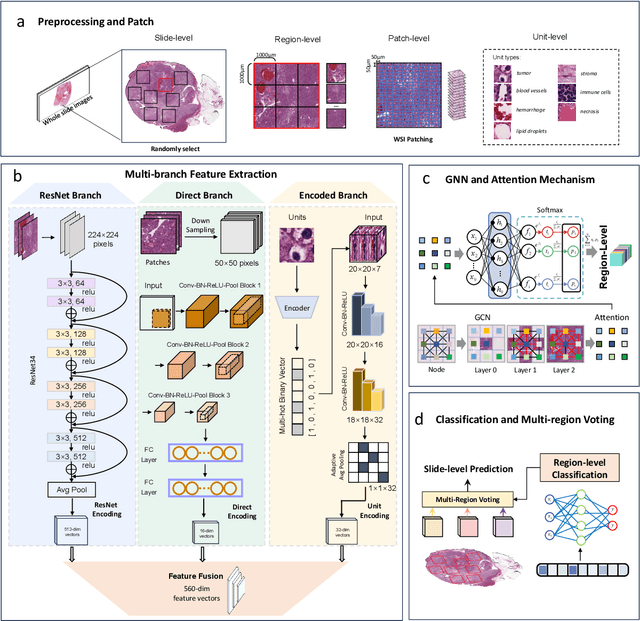
Abstract:Clear cell renal cell carcinoma (ccRCC) exhibits extensive intratumoral heterogeneity on multiple biological scales, contributing to variable clinical outcomes and limiting the effectiveness of conventional TNM staging, which highlights the urgent need for multiscale integrative analytic frameworks. The lipid-deficient de-clear cell differentiated (DCCD) ccRCC subtype, defined by multi-omics analyses, is associated with adverse outcomes even in early-stage disease. Here, we establish a hierarchical cross-scale framework for the preoperative identification of DCCD-ccRCC. At the highest layer, cross-modal mapping transferred molecular signatures to histological and CT phenotypes, establishing a molecular-to-pathology-to-radiology supervisory bridge. Within this framework, each modality-specific model is designed to mirror the inherent hierarchical structure of tumor biology. PathoDCCD captured multi-scale microscopic features, from cellular morphology and tissue architecture to meso-regional organization. RadioDCCD integrated complementary macroscopic information by combining whole-tumor and its habitat-subregions radiomics with a 2D maximal-section heterogeneity metric. These nested models enabled integrated molecular subtype prediction and clinical risk stratification. Across five cohorts totaling 1,659 patients, PathoDCCD reliably recapitulated molecular subtypes, while RadioDCCD provided reliable preoperative prediction. The consistent predictions identified patients with the poorest clinical outcomes. This cross-scale paradigm unifies molecular biology, computational pathology, and quantitative radiology into a biologically grounded strategy for preoperative noninvasive molecular phenotyping of ccRCC.
A hierarchical approach to deep learning and its application to tomographic reconstruction
Dec 16, 2019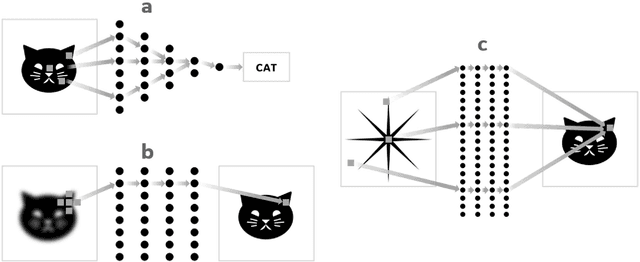
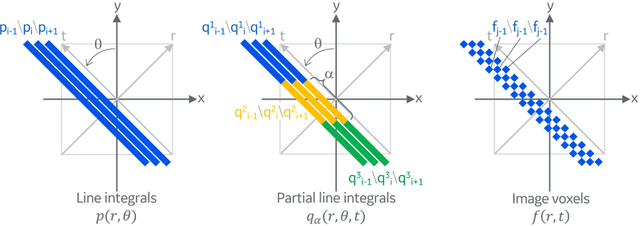
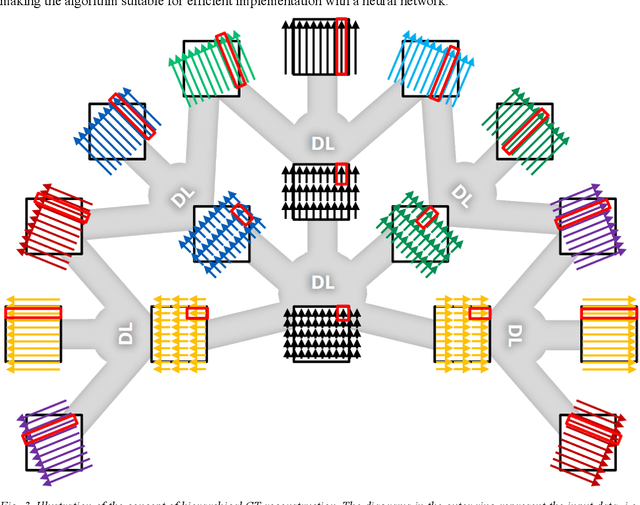
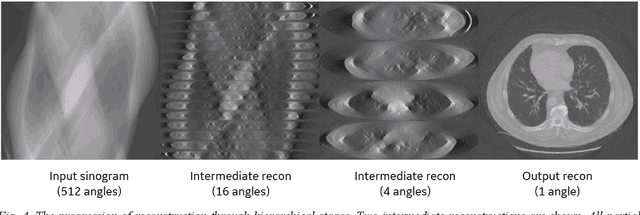
Abstract:Deep learning (DL) has shown unprecedented performance for many image analysis and image enhancement tasks. Yet, solving large-scale inverse problems like tomographic reconstruction remains challenging for DL. These problems involve non-local and space-variant integral transforms between the input and output domains, for which no efficient neural network models have been found. A prior attempt to solve such problems with supervised learning relied on a brute-force fully connected network and applied it to reconstruction for a $128^4$ system matrix size. This cannot practically scale to realistic data sizes such as $512^4$ and $512^6$ for three-dimensional data sets. Here we present a novel framework to solve such problems with deep learning by casting the original problem as a continuum of intermediate representations between the input and output data. The original problem is broken down into a sequence of simpler transformations that can be well mapped onto an efficient hierarchical network architecture, with exponentially fewer parameters than a generic network would need. We applied the approach to computed tomography (CT) image reconstruction for a $512^4$ system matrix size. To our knowledge, this enabled the first data-driven DL solver for full-size CT reconstruction without relying on the structure of direct (analytical) or iterative (numerical) inversion techniques. The proposed approach is applicable to other imaging problems such as emission and magnetic resonance reconstruction. More broadly, hierarchical DL opens the door to a new class of solvers for general inverse problems, which could potentially lead to improved signal-to-noise ratio, spatial resolution and computational efficiency in various areas.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge