Philip Naumann
Towards Robust Foundation Models for Digital Pathology
Jul 22, 2025
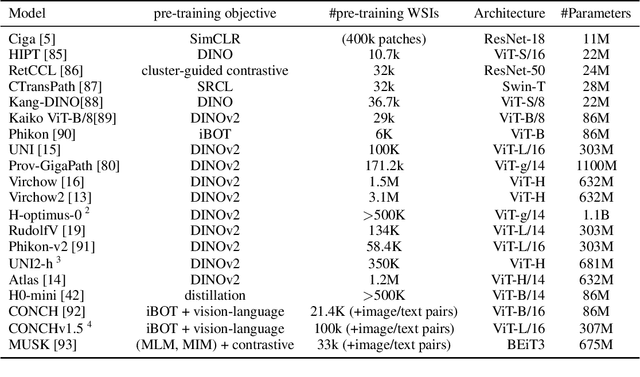
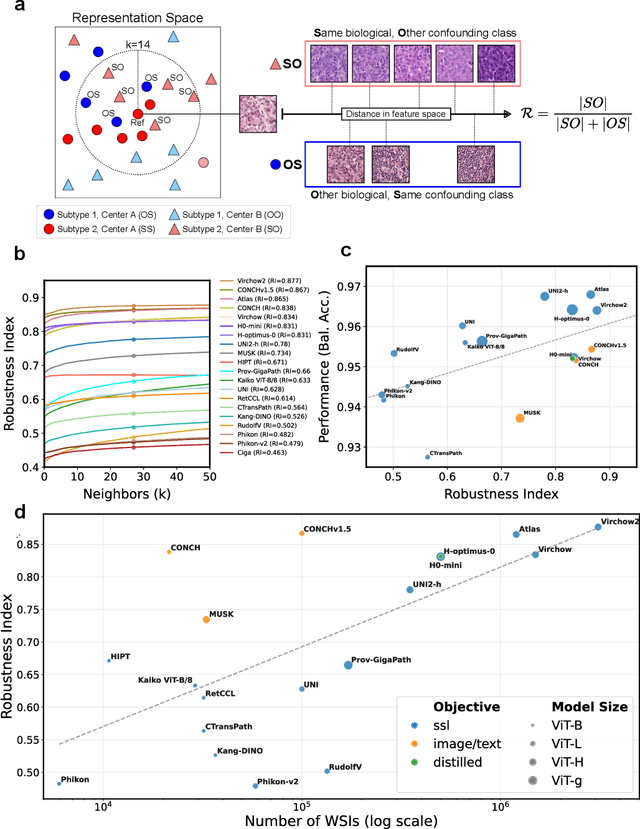

Abstract:Biomedical Foundation Models (FMs) are rapidly transforming AI-enabled healthcare research and entering clinical validation. However, their susceptibility to learning non-biological technical features -- including variations in surgical/endoscopic techniques, laboratory procedures, and scanner hardware -- poses risks for clinical deployment. We present the first systematic investigation of pathology FM robustness to non-biological features. Our work (i) introduces measures to quantify FM robustness, (ii) demonstrates the consequences of limited robustness, and (iii) proposes a framework for FM robustification to mitigate these issues. Specifically, we developed PathoROB, a robustness benchmark with three novel metrics, including the robustness index, and four datasets covering 28 biological classes from 34 medical centers. Our experiments reveal robustness deficits across all 20 evaluated FMs, and substantial robustness differences between them. We found that non-robust FM representations can cause major diagnostic downstream errors and clinical blunders that prevent safe clinical adoption. Using more robust FMs and post-hoc robustification considerably reduced (but did not yet eliminate) the risk of such errors. This work establishes that robustness evaluation is essential for validating pathology FMs before clinical adoption and demonstrates that future FM development must integrate robustness as a core design principle. PathoROB provides a blueprint for assessing robustness across biomedical domains, guiding FM improvement efforts towards more robust, representative, and clinically deployable AI systems that prioritize biological information over technical artifacts.
Wasserstein Distances Made Explainable: Insights into Dataset Shifts and Transport Phenomena
May 09, 2025Abstract:Wasserstein distances provide a powerful framework for comparing data distributions. They can be used to analyze processes over time or to detect inhomogeneities within data. However, simply calculating the Wasserstein distance or analyzing the corresponding transport map (or coupling) may not be sufficient for understanding what factors contribute to a high or low Wasserstein distance. In this work, we propose a novel solution based on Explainable AI that allows us to efficiently and accurately attribute Wasserstein distances to various data components, including data subgroups, input features, or interpretable subspaces. Our method achieves high accuracy across diverse datasets and Wasserstein distance specifications, and its practical utility is demonstrated in two use cases.
Consequence-aware Sequential Counterfactual Generation
Apr 12, 2021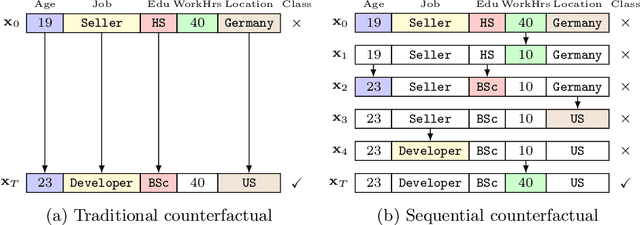
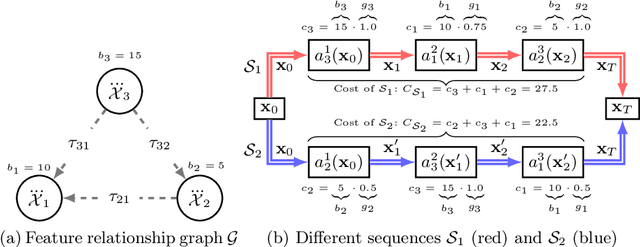
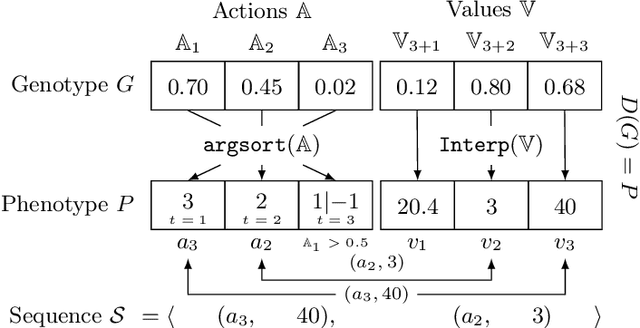
Abstract:Counterfactuals have become a popular technique nowadays for interacting with black-box machine learning models and understanding how to change a particular instance to obtain a desired outcome from the model. However, most existing approaches assume instant materialization of these changes, ignoring that they may require effort and a specific order of application. Recently, methods have been proposed that also consider the order in which actions are applied, leading to the so-called sequential counterfactual generation problem. In this work, we propose a model-agnostic method for sequential counterfactual generation. We formulate the task as a multi-objective optimization problem and present an evolutionary approach to find optimal sequences of actions leading to the counterfactuals. Our cost model considers not only the direct effect of an action, but also its consequences. Experimental results show that compared to state of the art, our approach generates less costly solutions, is more efficient, and provides the user with a diverse set of solutions to choose from.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge