Kathleen E. Larson
Self-Supervised CSF Inpainting with Synthetic Atrophy for Improved Accuracy Validation of Cortical Surface Analyses
Mar 10, 2023



Abstract:Accuracy validation of cortical thickness measurement is a difficult problem due to the lack of ground truth data. To address this need, many methods have been developed to synthetically induce gray matter (GM) atrophy in an MRI via deformable registration, creating a set of images with known changes in cortical thickness. However, these methods often cause blurring in atrophied regions, and cannot simulate realistic atrophy within deep sulci where cerebrospinal fluid (CSF) is obscured or absent. In this paper, we present a solution using a self-supervised inpainting model to generate CSF in these regions and create images with more plausible GM/CSF boundaries. Specifically, we introduce a novel, 3D GAN model that incorporates patch-based dropout training, edge map priors, and sinusoidal positional encoding, all of which are established methods previously limited to 2D domains. We show that our framework significantly improves the quality of the resulting synthetic images and is adaptable to unseen data with fine-tuning. We also demonstrate that our resulting dataset can be employed for accuracy validation of cortical segmentation and thickness measurement.
Unsupervised Cross-Modality Domain Adaptation for Segmenting Vestibular Schwannoma and Cochlea with Data Augmentation and Model Ensemble
Sep 24, 2021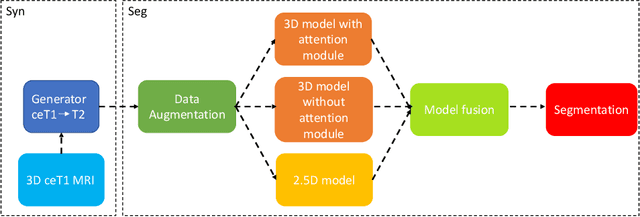
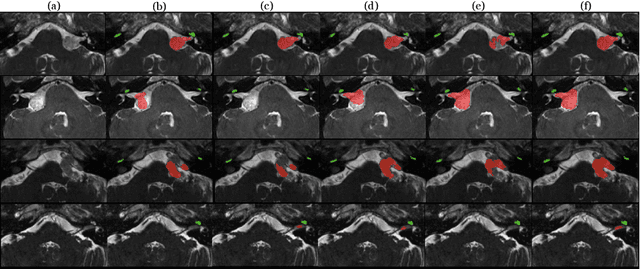
Abstract:Magnetic resonance images (MRIs) are widely used to quantify vestibular schwannoma and the cochlea. Recently, deep learning methods have shown state-of-the-art performance for segmenting these structures. However, training segmentation models may require manual labels in target domain, which is expensive and time-consuming. To overcome this problem, domain adaptation is an effective way to leverage information from source domain to obtain accurate segmentations without requiring manual labels in target domain. In this paper, we propose an unsupervised learning framework to segment the VS and cochlea. Our framework leverages information from contrast-enhanced T1-weighted (ceT1-w) MRIs and its labels, and produces segmentations for T2-weighted MRIs without any labels in the target domain. We first applied a generator to achieve image-to-image translation. Next, we ensemble outputs from an ensemble of different models to obtain final segmentations. To cope with MRIs from different sites/scanners, we applied various 'online' augmentations during training to better capture the geometric variability and the variability in image appearance and quality. Our method is easy to build and produces promising segmentations, with a mean Dice score of 0.7930 and 0.7432 for VS and cochlea respectively in the validation set.
LIFE: A Generalizable Autodidactic Pipeline for 3D OCT-A Vessel Segmentation
Jul 09, 2021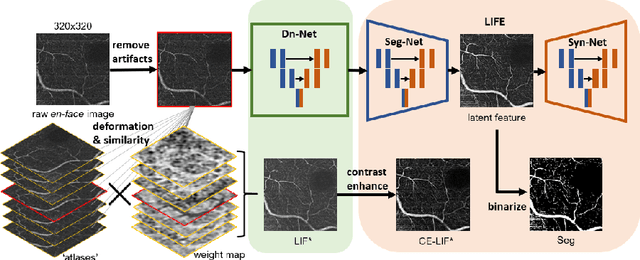
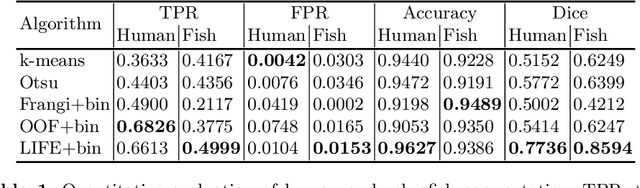
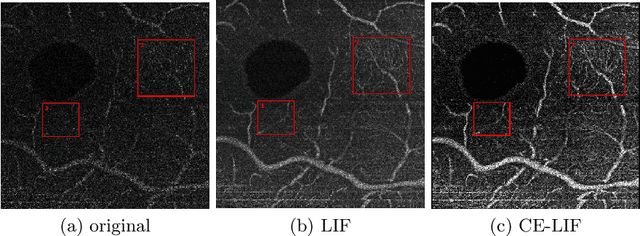
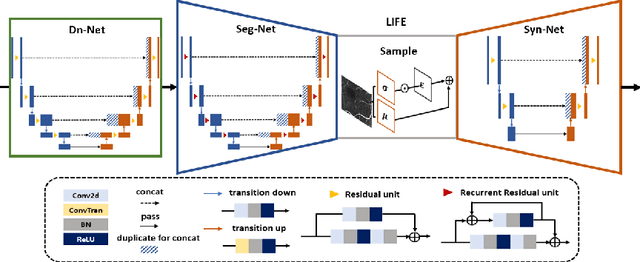
Abstract:Optical coherence tomography (OCT) is a non-invasive imaging technique widely used for ophthalmology. It can be extended to OCT angiography (OCT-A), which reveals the retinal vasculature with improved contrast. Recent deep learning algorithms produced promising vascular segmentation results; however, 3D retinal vessel segmentation remains difficult due to the lack of manually annotated training data. We propose a learning-based method that is only supervised by a self-synthesized modality named local intensity fusion (LIF). LIF is a capillary-enhanced volume computed directly from the input OCT-A. We then construct the local intensity fusion encoder (LIFE) to map a given OCT-A volume and its LIF counterpart to a shared latent space. The latent space of LIFE has the same dimensions as the input data and it contains features common to both modalities. By binarizing this latent space, we obtain a volumetric vessel segmentation. Our method is evaluated in a human fovea OCT-A and three zebrafish OCT-A volumes with manual labels. It yields a Dice score of 0.7736 on human data and 0.8594 +/- 0.0275 on zebrafish data, a dramatic improvement over existing unsupervised algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge