Karl Deisseroth
POCO: Scalable Neural Forecasting through Population Conditioning
Jun 17, 2025Abstract:Predicting future neural activity is a core challenge in modeling brain dynamics, with applications ranging from scientific investigation to closed-loop neurotechnology. While recent models of population activity emphasize interpretability and behavioral decoding, neural forecasting-particularly across multi-session, spontaneous recordings-remains underexplored. We introduce POCO, a unified forecasting model that combines a lightweight univariate forecaster with a population-level encoder to capture both neuron-specific and brain-wide dynamics. Trained across five calcium imaging datasets spanning zebrafish, mice, and C. elegans, POCO achieves state-of-the-art accuracy at cellular resolution in spontaneous behaviors. After pre-training, POCO rapidly adapts to new recordings with minimal fine-tuning. Notably, POCO's learned unit embeddings recover biologically meaningful structure-such as brain region clustering-without any anatomical labels. Our comprehensive analysis reveals several key factors influencing performance, including context length, session diversity, and preprocessing. Together, these results position POCO as a scalable and adaptable approach for cross-session neural forecasting and offer actionable insights for future model design. By enabling accurate, generalizable forecasting models of neural dynamics across individuals and species, POCO lays the groundwork for adaptive neurotechnologies and large-scale efforts for neural foundation models.
A Large Deformation Diffeomorphic Approach to Registration of CLARITY Images via Mutual Information
Aug 11, 2017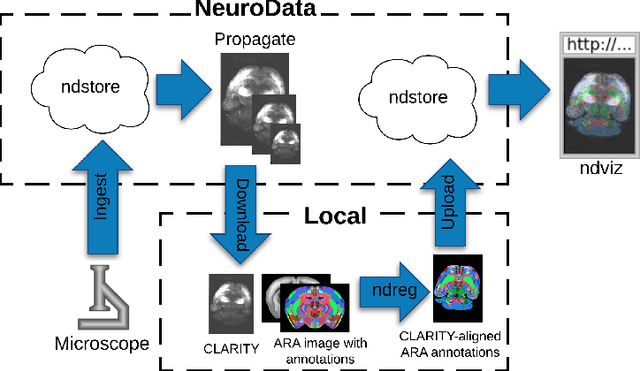
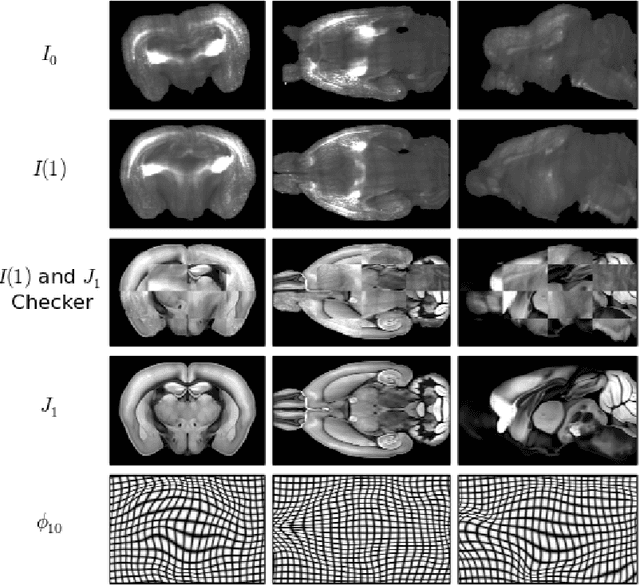
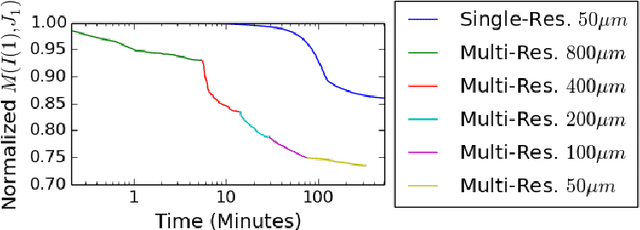
Abstract:CLARITY is a method for converting biological tissues into translucent and porous hydrogel-tissue hybrids. This facilitates interrogation with light sheet microscopy and penetration of molecular probes while avoiding physical slicing. In this work, we develop a pipeline for registering CLARIfied mouse brains to an annotated brain atlas. Due to the novelty of this microscopy technique it is impractical to use absolute intensity values to align these images to existing standard atlases. Thus we adopt a large deformation diffeomorphic approach for registering images via mutual information matching. Furthermore we show how a cascaded multi-resolution approach can improve registration quality while reducing algorithm run time. As acquired image volumes were over a terabyte in size, they were far too large for work on personal computers. Therefore the NeuroData computational infrastructure was deployed for multi-resolution storage and visualization of these images and aligned annotations on the web.
Deformably Registering and Annotating Whole CLARITY Brains to an Atlas via Masked LDDMM
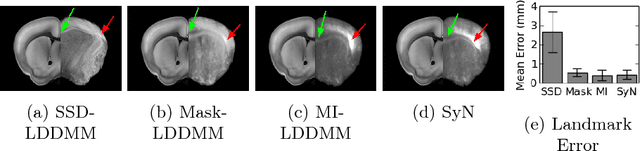
May 06, 2016Abstract:The CLARITY method renders brains optically transparent to enable high-resolution imaging in the structurally intact brain. Anatomically annotating CLARITY brains is necessary for discovering which regions contain signals of interest. Manually annotating whole-brain, terabyte CLARITY images is difficult, time-consuming, subjective, and error-prone. Automatically registering CLARITY images to a pre-annotated brain atlas offers a solution, but is difficult for several reasons. Removal of the brain from the skull and subsequent storage and processing cause variable non-rigid deformations, thus compounding inter-subject anatomical variability. Additionally, the signal in CLARITY images arises from various biochemical contrast agents which only sparsely label brain structures. This sparse labeling challenges the most commonly used registration algorithms that need to match image histogram statistics to the more densely labeled histological brain atlases. The standard method is a multiscale Mutual Information B-spline algorithm that dynamically generates an average template as an intermediate registration target. We determined that this method performs poorly when registering CLARITY brains to the Allen Institute's Mouse Reference Atlas (ARA), because the image histogram statistics are poorly matched. Therefore, we developed a method (Mask-LDDMM) for registering CLARITY images, that automatically find the brain boundary and learns the optimal deformation between the brain and atlas masks. Using Mask-LDDMM without an average template provided better results than the standard approach when registering CLARITY brains to the ARA. The LDDMM pipelines developed here provide a fast automated way to anatomically annotate CLARITY images. Our code is available as open source software at http://NeuroData.io.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge