Juan Xie
Robust Ultra-High-Dimensional Variable Selection With Correlated Structure Using Group Testing
Feb 06, 2026Abstract:Background: High-dimensional genomic data exhibit strong group correlation structures that challenge conventional feature selection methods, which often assume feature independence or rely on pre-defined pathways and are sensitive to outliers and model misspecification. Methods: We propose the Dorfman screening framework, a multi-stage procedure that forms data-driven variable groups via hierarchical clustering, performs group and within-group hypothesis testing, and refines selection using elastic net or adaptive elastic net. Robust variants incorporate OGK-based covariance estimation, rank-based correlation, and Huber-weighted regression to handle contaminated and non-normal data. Results: In simulations, Dorfman-Sparse-Adaptive-EN performed best under normal conditions, while Robust-OGK-Dorfman-Adaptive-EN showed clear advantages under data contamination, outperforming classical Dorfman and competing methods. Applied to NSCLC gene expression data for trametinib response, robust Dorfman methods achieved the lowest prediction errors and enriched recovery of clinically relevant genes. Conclusions: The Dorfman framework provides an efficient and robust approach to genomic feature selection. Robust-OGK-Dorfman-Adaptive-EN offers strong performance under both ideal and contaminated conditions and scales to ultra-high-dimensional settings, making it well suited for modern genomic biomarker discovery.
Source-free Active Domain Adaptation for Diabetic Retinopathy Grading Based on Ultra-wide-field Fundus Image
Sep 19, 2023Abstract:Domain adaptation (DA) has been widely applied in the diabetic retinopathy (DR) grading of unannotated ultra-wide-field (UWF) fundus images, which can transfer annotated knowledge from labeled color fundus images. However, suffering from huge domain gaps and complex real-world scenarios, the DR grading performance of most mainstream DA is far from that of clinical diagnosis. To tackle this, we propose a novel source-free active domain adaptation (SFADA) in this paper. Specifically, we focus on DR grading problem itself and propose to generate features of color fundus images with continuously evolving relationships of DRs, actively select a few valuable UWF fundus images for labeling with local representation matching, and adapt model on UWF fundus images with DR lesion prototypes. Notably, the SFADA also takes data privacy and computational efficiency into consideration. Extensive experimental results demonstrate that our proposed SFADA achieves state-of-the-art DR grading performance, increasing accuracy by 20.9% and quadratic weighted kappa by 18.63% compared with baseline and reaching 85.36% and 92.38% respectively. These investigations show that the potential of our approach for real clinical practice is promising.
A comprehensive review of 3D point cloud descriptors
Feb 07, 2018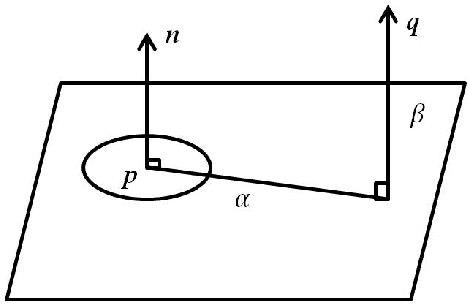
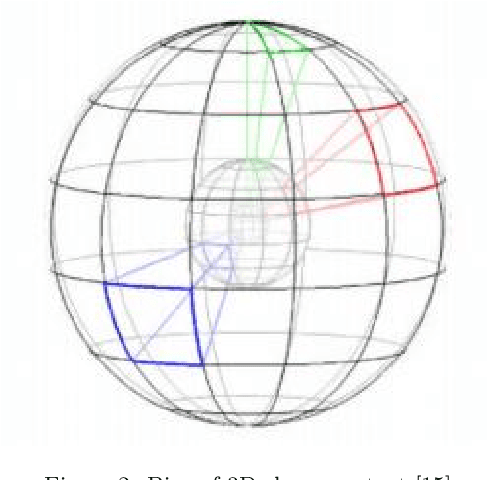
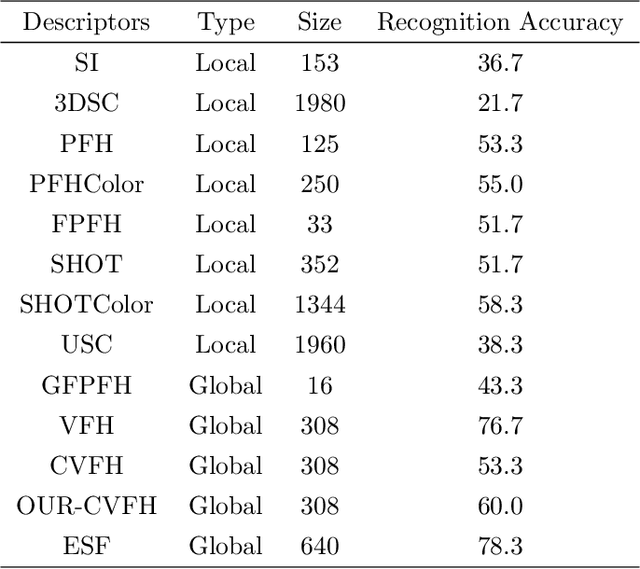
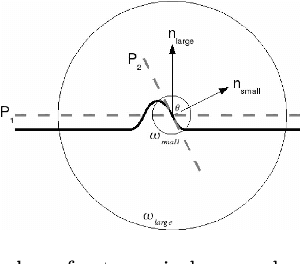
Abstract:The introduction of inexpensive 3D data acquisition devices has promisingly facilitated the wide availability and popularity of 3D point cloud, which attracts more attention on the effective extraction of novel 3D point cloud descriptors for accurate and efficient of 3D computer vision tasks. However, how to de- velop discriminative and robust feature descriptors from various point clouds remains a challenging task. This paper comprehensively investigates the exist- ing approaches for extracting 3D point cloud descriptors which are categorized into three major classes: local-based descriptor, global-based descriptor and hybrid-based descriptor. Furthermore, experiments are carried out to present a thorough evaluation of performance of several state-of-the-art 3D point cloud descriptors used widely in practice in terms of descriptiveness, robustness and efficiency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge