Joshua Robinson
Michael Pokorny
Sign and Basis Invariant Networks for Spectral Graph Representation Learning
Apr 11, 2022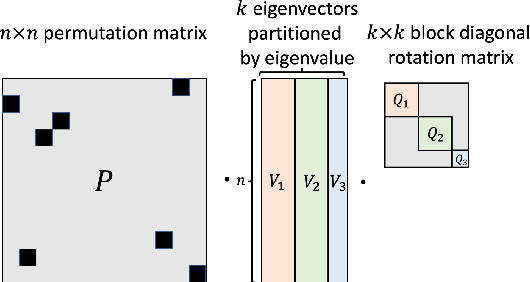
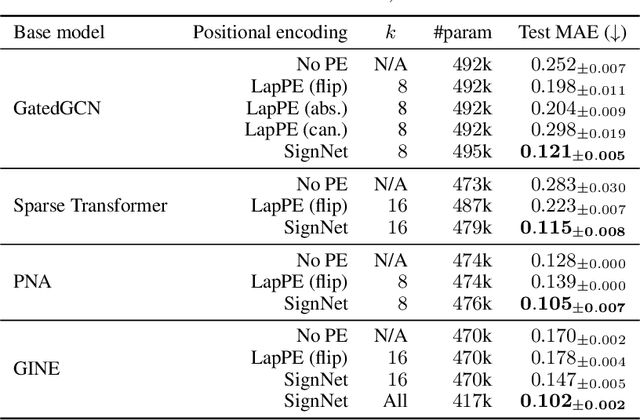
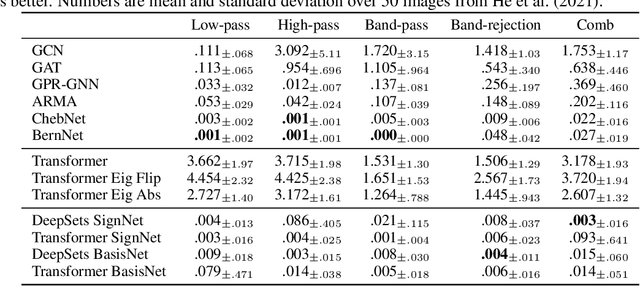
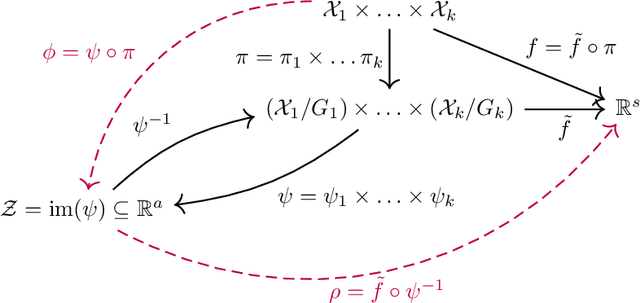
Abstract:Many machine learning tasks involve processing eigenvectors derived from data. Especially valuable are Laplacian eigenvectors, which capture useful structural information about graphs and other geometric objects. However, ambiguities arise when computing eigenvectors: for each eigenvector $v$, the sign flipped $-v$ is also an eigenvector. More generally, higher dimensional eigenspaces contain infinitely many choices of basis eigenvectors. These ambiguities make it a challenge to process eigenvectors and eigenspaces in a consistent way. In this work we introduce SignNet and BasisNet -- new neural architectures that are invariant to all requisite symmetries and hence process collections of eigenspaces in a principled manner. Our networks are universal, i.e., they can approximate any continuous function of eigenvectors with the proper invariances. They are also theoretically strong for graph representation learning -- they can approximate any spectral graph convolution, can compute spectral invariants that go beyond message passing neural networks, and can provably simulate previously proposed graph positional encodings. Experiments show the strength of our networks for molecular graph regression, learning expressive graph representations, and learning implicit neural representations on triangle meshes. Our code is available at https://github.com/cptq/SignNet-BasisNet .
An Information-theoretic Approach to Prompt Engineering Without Ground Truth Labels
Mar 21, 2022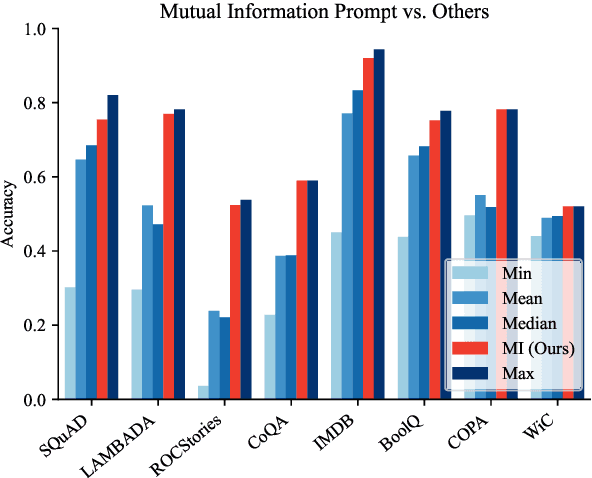
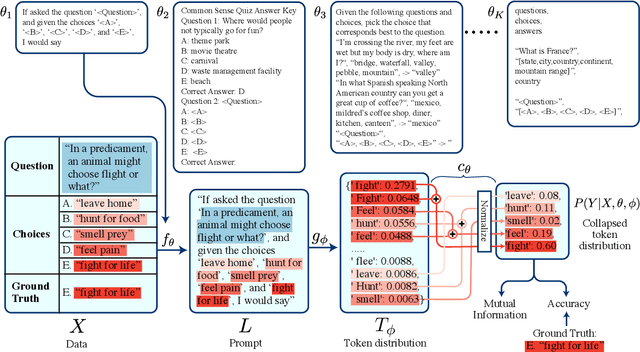
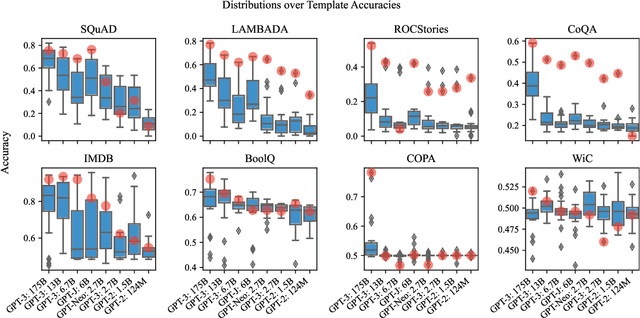
Abstract:Pre-trained language models derive substantial linguistic and factual knowledge from the massive corpora on which they are trained, and prompt engineering seeks to align these models to specific tasks. Unfortunately, existing prompt engineering methods require significant amounts of labeled data, access to model parameters, or both. We introduce a new method for selecting prompt templates \textit{without labeled examples} and \textit{without direct access to the model}. Specifically, over a set of candidate templates, we choose the template that maximizes the mutual information between the input and the corresponding model output. Across 8 datasets representing 7 distinct NLP tasks, we show that when a template has high mutual information, it also has high accuracy on the task. On the largest model, selecting prompts with our method gets 90\% of the way from the average prompt accuracy to the best prompt accuracy and requires no ground truth labels.
Can contrastive learning avoid shortcut solutions?
Jun 21, 2021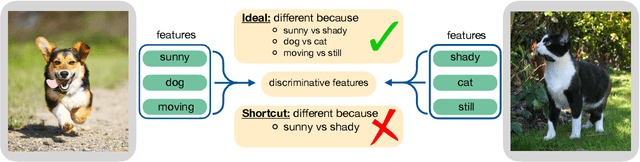
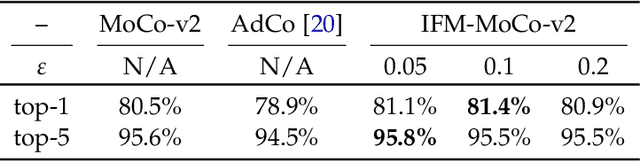
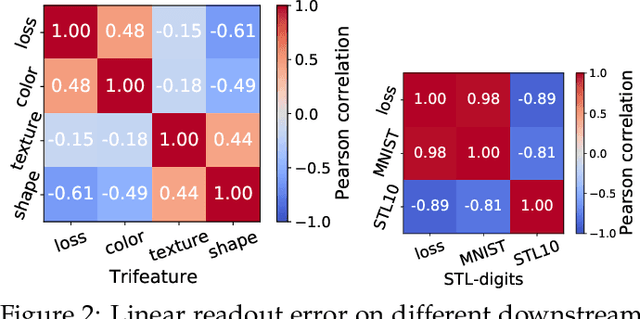
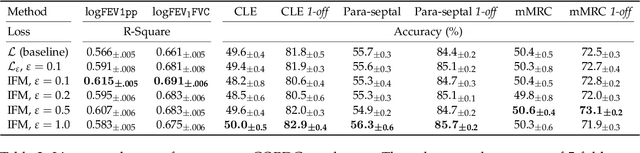
Abstract:The generalization of representations learned via contrastive learning depends crucially on what features of the data are extracted. However, we observe that the contrastive loss does not always sufficiently guide which features are extracted, a behavior that can negatively impact the performance on downstream tasks via "shortcuts", i.e., by inadvertently suppressing important predictive features. We find that feature extraction is influenced by the difficulty of the so-called instance discrimination task (i.e., the task of discriminating pairs of similar points from pairs of dissimilar ones). Although harder pairs improve the representation of some features, the improvement comes at the cost of suppressing previously well represented features. In response, we propose implicit feature modification (IFM), a method for altering positive and negative samples in order to guide contrastive models towards capturing a wider variety of predictive features. Empirically, we observe that IFM reduces feature suppression, and as a result improves performance on vision and medical imaging tasks. The code is available at: \url{https://github.com/joshr17/IFM}.
A Matrix Autoencoder Framework to Align the Functional and Structural Connectivity Manifolds as Guided by Behavioral Phenotypes
May 30, 2021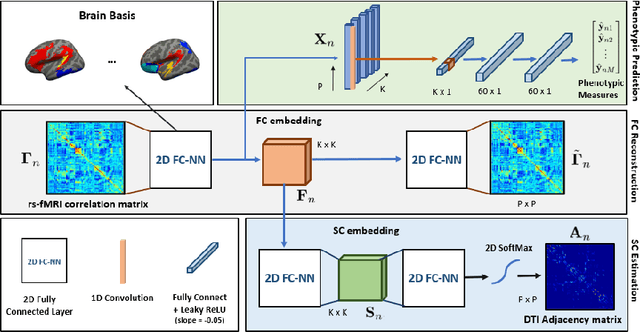
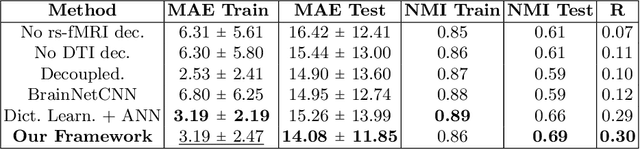
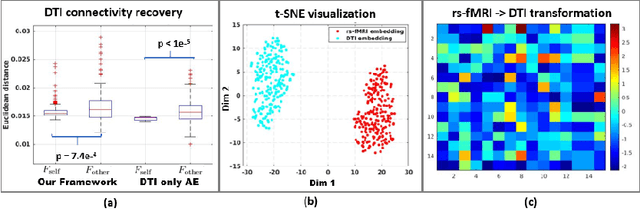
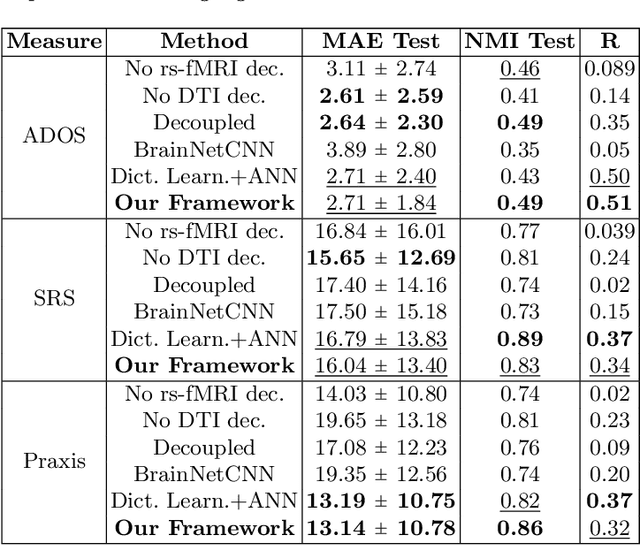
Abstract:We propose a novel matrix autoencoder to map functional connectomes from resting state fMRI (rs-fMRI) to structural connectomes from Diffusion Tensor Imaging (DTI), as guided by subject-level phenotypic measures. Our specialized autoencoder infers a low dimensional manifold embedding for the rs-fMRI correlation matrices that mimics a canonical outer-product decomposition. The embedding is simultaneously used to reconstruct DTI tractography matrices via a second manifold alignment decoder and to predict inter-subject phenotypic variability via an artificial neural network. We validate our framework on a dataset of 275 healthy individuals from the Human Connectome Project database and on a second clinical dataset consisting of 57 subjects with Autism Spectrum Disorder. We demonstrate that the model reliably recovers structural connectivity patterns across individuals, while robustly extracting predictive and interpretable brain biomarkers in a cross-validated setting. Finally, our framework outperforms several baselines at predicting behavioral phenotypes in both real-world datasets.
Contrastive Learning with Hard Negative Samples
Oct 09, 2020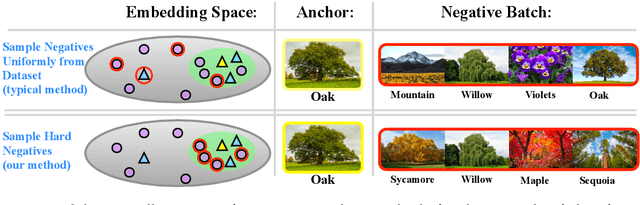
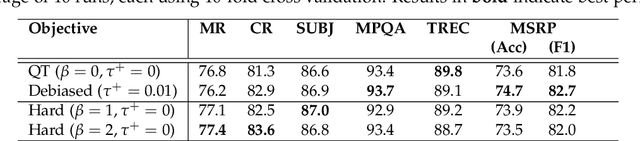
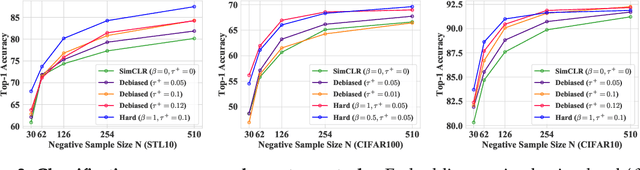
Abstract:We consider the question: how can you sample good negative examples for contrastive learning? We argue that, as with metric learning, learning contrastive representations benefits from hard negative samples (i.e., points that are difficult to distinguish from an anchor point). The key challenge toward using hard negatives is that contrastive methods must remain unsupervised, making it infeasible to adopt existing negative sampling strategies that use label information. In response, we develop a new class of unsupervised methods for selecting hard negative samples where the user can control the amount of hardness. A limiting case of this sampling results in a representation that tightly clusters each class, and pushes different classes as far apart as possible. The proposed method improves downstream performance across multiple modalities, requires only few additional lines of code to implement, and introduces no computational overhead.
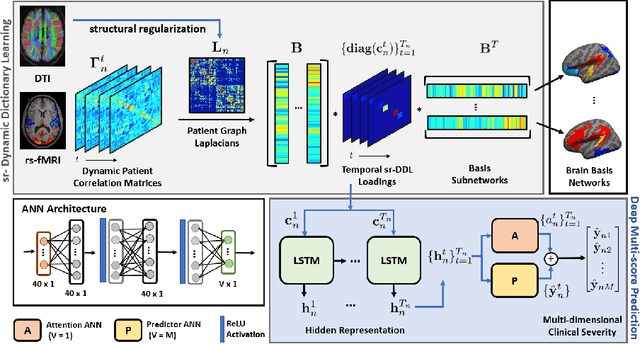
Deep sr-DDL: Deep Structurally Regularized Dynamic Dictionary Learning to Integrate Multimodal and Dynamic Functional Connectomics data for Multidimensional Clinical Characterizations
Aug 27, 2020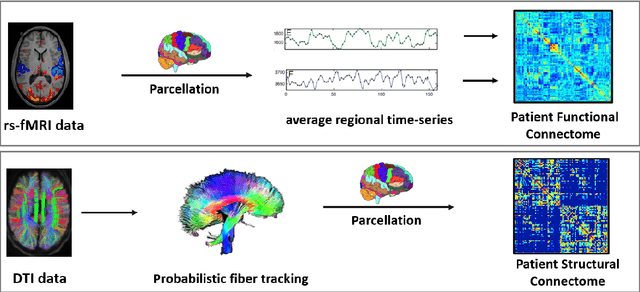
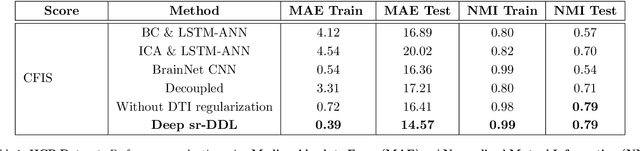
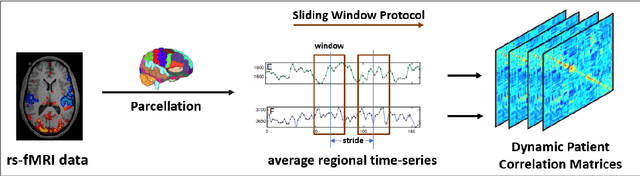
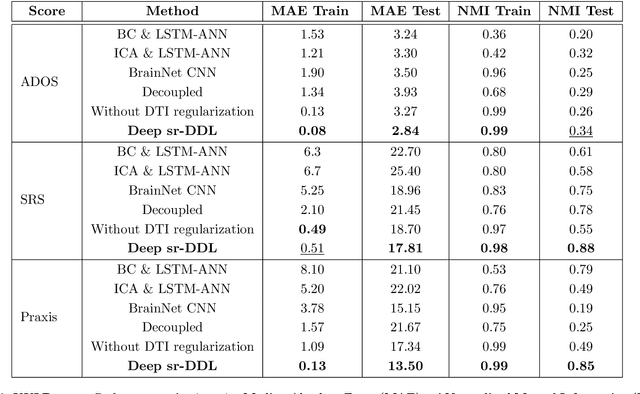
Abstract:We propose a novel integrated framework that jointly models complementary information from resting-state functional MRI (rs-fMRI) connectivity and diffusion tensor imaging (DTI) tractography to extract biomarkers of brain connectivity predictive of behavior. Our framework couples a generative model of the connectomics data with a deep network that predicts behavioral scores. The generative component is a structurally-regularized Dynamic Dictionary Learning (sr-DDL) model that decomposes the dynamic rs-fMRI correlation matrices into a collection of shared basis networks and time varying subject-specific loadings. We use the DTI tractography to regularize this matrix factorization and learn anatomically informed functional connectivity profiles. The deep component of our framework is an LSTM-ANN block, which uses the temporal evolution of the subject-specific sr-DDL loadings to predict multidimensional clinical characterizations. Our joint optimization strategy collectively estimates the basis networks, the subject-specific time-varying loadings, and the neural network weights. We validate our framework on a dataset of neurotypical individuals from the Human Connectome Project (HCP) database to map to cognition and on a separate multi-score prediction task on individuals diagnosed with Autism Spectrum Disorder (ASD) in a five-fold cross validation setting. Our hybrid model outperforms several state-of-the-art approaches at clinical outcome prediction and learns interpretable multimodal neural signatures of brain organization.
Debiased Contrastive Learning
Jul 05, 2020
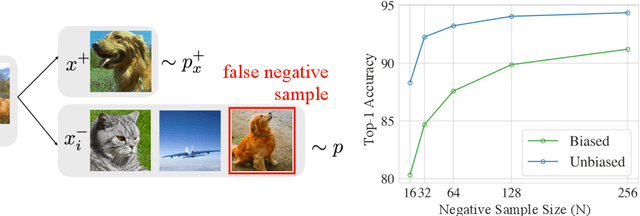
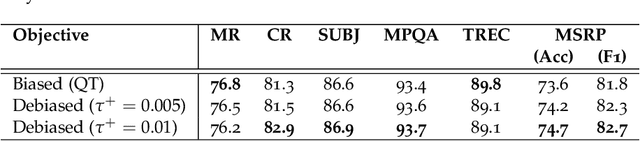

Abstract:A prominent technique for self-supervised representation learning has been to contrast semantically similar and dissimilar pairs of samples. Without access to labels, dissimilar (negative) points are typically taken to be randomly sampled datapoints, implicitly accepting that these points may, in reality, actually have the same label. Perhaps unsurprisingly, we observe that sampling negative examples from truly different labels improves performance, in a synthetic setting where labels are available. Motivated by this observation, we develop a debiased contrastive objective that corrects for the sampling of same-label datapoints, even without knowledge of the true labels. Empirically, the proposed objective consistently outperforms the state-of-the-art for representation learning in vision, language, and reinforcement learning benchmarks. Theoretically, we establish generalization bounds for the downstream classification task.
A Deep-Generative Hybrid Model to Integrate Multimodal and Dynamic Connectivity for Predicting Spectrum-Level Deficits in Autism
Jul 03, 2020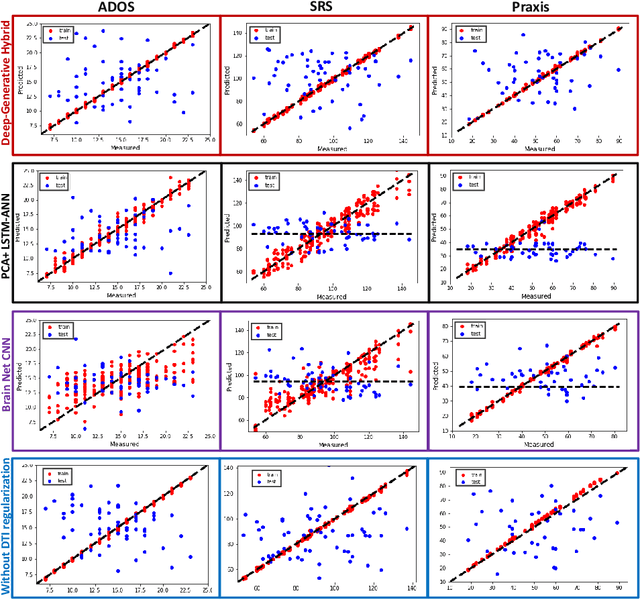
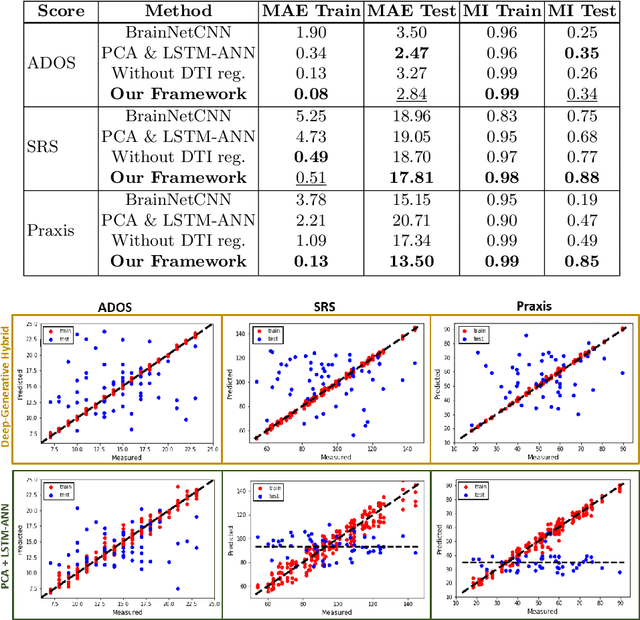
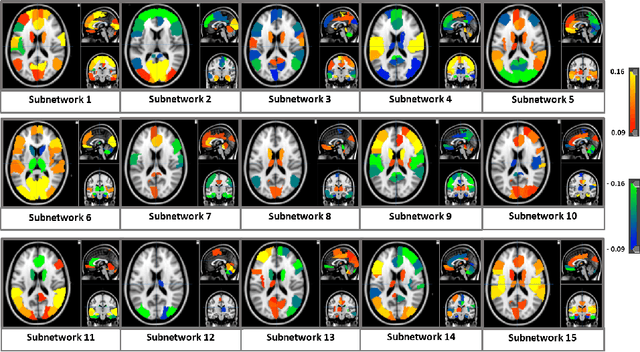
Abstract:We propose an integrated deep-generative framework, that jointly models complementary information from resting-state functional MRI (rs-fMRI) connectivity and diffusion tensor imaging (DTI) tractography to extract predictive biomarkers of a disease. The generative part of our framework is a structurally-regularized Dynamic Dictionary Learning (sr-DDL) model that decomposes the dynamic rs-fMRI correlation matrices into a collection of shared basis networks and time varying patient-specific loadings. This matrix factorization is guided by the DTI tractography matrices to learn anatomically informed connectivity profiles. The deep part of our framework is an LSTM-ANN block, which models the temporal evolution of the patient sr-DDL loadings to predict multidimensional clinical severity. Our coupled optimization procedure collectively estimates the basis networks, the patient-specific dynamic loadings, and the neural network weights. We validate our framework on a multi-score prediction task in 57 patients diagnosed with Autism Spectrum Disorder (ASD). Our hybrid model outperforms state-of-the-art baselines in a five-fold cross validated setting and extracts interpretable multimodal neural signatures of brain dysfunction in ASD.
Strength from Weakness: Fast Learning Using Weak Supervision
Feb 19, 2020
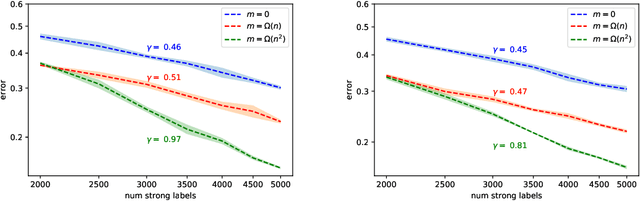
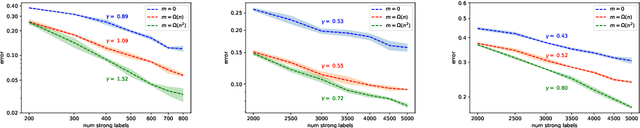
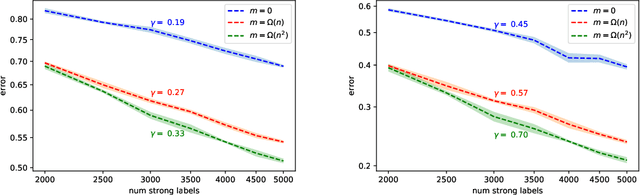
Abstract:We study generalization properties of weakly supervised learning. That is, learning where only a few "strong" labels (the actual target of our prediction) are present but many more "weak" labels are available. In particular, we show that having access to weak labels can significantly accelerate the learning rate for the strong task to the fast rate of $\mathcal{O}(\nicefrac1n)$, where $n$ denotes the number of strongly labeled data points. This acceleration can happen even if by itself the strongly labeled data admits only the slower $\mathcal{O}(\nicefrac{1}{\sqrt{n}})$ rate. The actual acceleration depends continuously on the number of weak labels available, and on the relation between the two tasks. Our theoretical results are reflected empirically across a range of tasks and illustrate how weak labels speed up learning on the strong task.
Flexible Modeling of Diversity with Strongly Log-Concave Distributions
Jun 12, 2019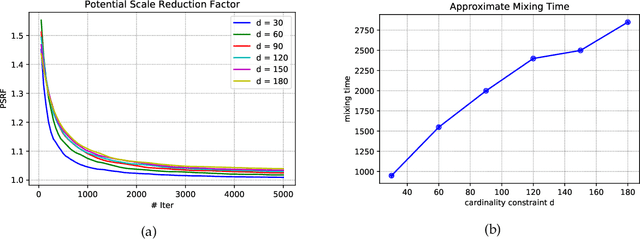
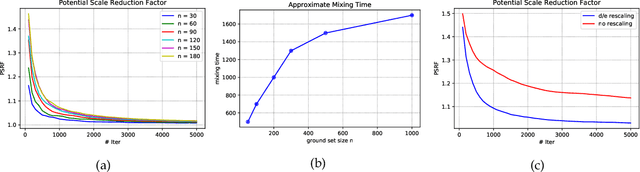
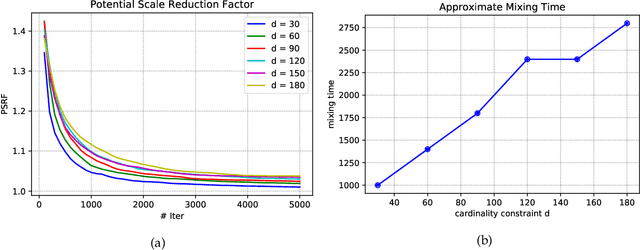
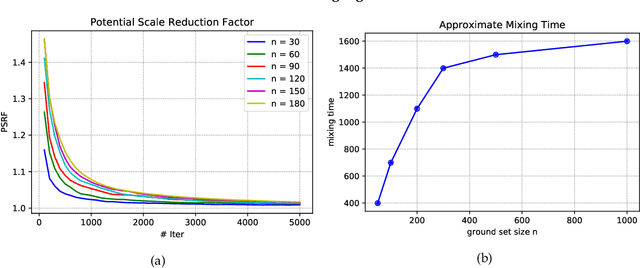
Abstract:Strongly log-concave (SLC) distributions are a rich class of discrete probability distributions over subsets of some ground set. They are strictly more general than strongly Rayleigh (SR) distributions such as the well-known determinantal point process. While SR distributions offer elegant models of diversity, they lack an easy control over how they express diversity. We propose SLC as the right extension of SR that enables easier, more intuitive control over diversity, illustrating this via examples of practical importance. We develop two fundamental tools needed to apply SLC distributions to learning and inference: sampling and mode finding. For sampling we develop an MCMC sampler and give theoretical mixing time bounds. For mode finding, we establish a weak log-submodularity property for SLC functions and derive optimization guarantees for a distorted greedy algorithm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge