Joseph Lo
Learning to Diagnose Privately: DP-Powered LLMs for Radiology Report Classification
Jun 04, 2025Abstract:Purpose: This study proposes a framework for fine-tuning large language models (LLMs) with differential privacy (DP) to perform multi-abnormality classification on radiology report text. By injecting calibrated noise during fine-tuning, the framework seeks to mitigate the privacy risks associated with sensitive patient data and protect against data leakage while maintaining classification performance. Materials and Methods: We used 50,232 radiology reports from the publicly available MIMIC-CXR chest radiography and CT-RATE computed tomography datasets, collected between 2011 and 2019. Fine-tuning of LLMs was conducted to classify 14 labels from MIMIC-CXR dataset, and 18 labels from CT-RATE dataset using Differentially Private Low-Rank Adaptation (DP-LoRA) in high and moderate privacy regimes (across a range of privacy budgets = {0.01, 0.1, 1.0, 10.0}). Model performance was evaluated using weighted F1 score across three model architectures: BERT-medium, BERT-small, and ALBERT-base. Statistical analyses compared model performance across different privacy levels to quantify the privacy-utility trade-off. Results: We observe a clear privacy-utility trade-off through our experiments on 2 different datasets and 3 different models. Under moderate privacy guarantees the DP fine-tuned models achieved comparable weighted F1 scores of 0.88 on MIMIC-CXR and 0.59 on CT-RATE, compared to non-private LoRA baselines of 0.90 and 0.78, respectively. Conclusion: Differentially private fine-tuning using LoRA enables effective and privacy-preserving multi-abnormality classification from radiology reports, addressing a key challenge in fine-tuning LLMs on sensitive medical data.
XCAT-2.0: A Comprehensive Library of Personalized Digital Twins Derived from CT Scans
May 18, 2024



Abstract:Virtual Imaging Trials (VIT) offer a cost-effective and scalable approach for evaluating medical imaging technologies. Computational phantoms, which mimic real patient anatomy and physiology, play a central role in VIT. However, the current libraries of computational phantoms face limitations, particularly in terms of sample size and diversity. Insufficient representation of the population hampers accurate assessment of imaging technologies across different patient groups. Traditionally, phantoms were created by manual segmentation, which is a laborious and time-consuming task, impeding the expansion of phantom libraries. This study presents a framework for realistic computational phantom modeling using a suite of four deep learning segmentation models, followed by three forms of automated organ segmentation quality control. Over 2500 computational phantoms with up to 140 structures illustrating a sophisticated approach to detailed anatomical modeling are released. Phantoms are available in both voxelized and surface mesh formats. The framework is aggregated with an in-house CT scanner simulator to produce realistic CT images. The framework can potentially advance virtual imaging trials, facilitating comprehensive and reliable evaluations of medical imaging technologies. Phantoms may be requested at https://cvit.duke.edu/resources/, code, model weights, and sample CT images are available at https://xcat-2.github.io.
Mask Embedding in conditional GAN for Guided Synthesis of High Resolution Images
Jul 03, 2019

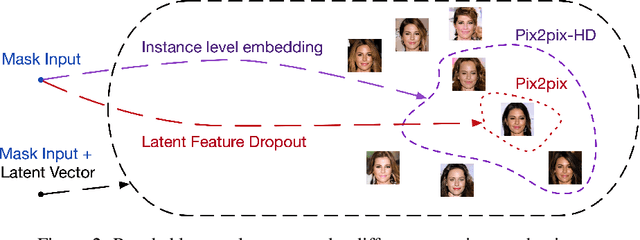
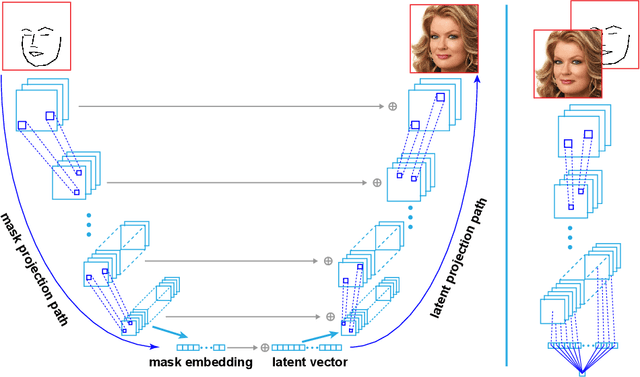
Abstract:Recent advancements in conditional Generative Adversarial Networks (cGANs) have shown promises in label guided image synthesis. Semantic masks, such as sketches and label maps, are another intuitive and effective form of guidance in image synthesis. Directly incorporating the semantic masks as constraints dramatically reduces the variability and quality of the synthesized results. We observe this is caused by the incompatibility of features from different inputs (such as mask image and latent vector) of the generator. To use semantic masks as guidance whilst providing realistic synthesized results with fine details, we propose to use mask embedding mechanism to allow for a more efficient initial feature projection in the generator. We validate the effectiveness of our approach by training a mask guided face generator using CELEBA-HQ dataset. We can generate realistic and high resolution facial images up to the resolution of 512*512 with a mask guidance. Our code is publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge