Jigang Fan
Evolutionary Profiles for Protein Fitness Prediction
Oct 08, 2025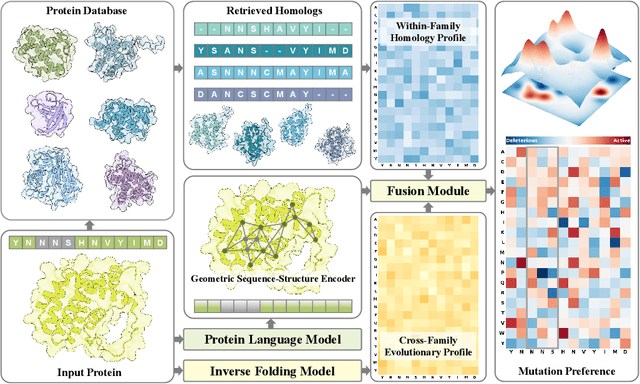
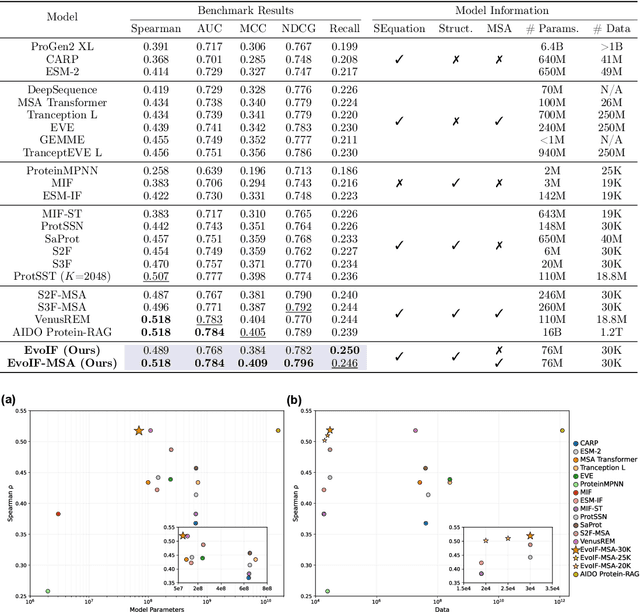
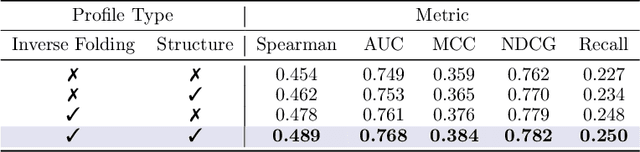
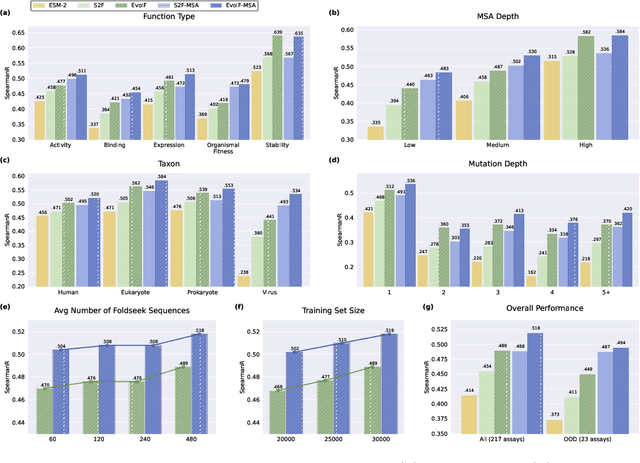
Abstract:Predicting the fitness impact of mutations is central to protein engineering but constrained by limited assays relative to the size of sequence space. Protein language models (pLMs) trained with masked language modeling (MLM) exhibit strong zero-shot fitness prediction; we provide a unifying view by interpreting natural evolution as implicit reward maximization and MLM as inverse reinforcement learning (IRL), in which extant sequences act as expert demonstrations and pLM log-odds serve as fitness estimates. Building on this perspective, we introduce EvoIF, a lightweight model that integrates two complementary sources of evolutionary signal: (i) within-family profiles from retrieved homologs and (ii) cross-family structural-evolutionary constraints distilled from inverse folding logits. EvoIF fuses sequence-structure representations with these profiles via a compact transition block, yielding calibrated probabilities for log-odds scoring. On ProteinGym (217 mutational assays; >2.5M mutants), EvoIF and its MSA-enabled variant achieve state-of-the-art or competitive performance while using only 0.15% of the training data and fewer parameters than recent large models. Ablations confirm that within-family and cross-family profiles are complementary, improving robustness across function types, MSA depths, taxa, and mutation depths. The codes will be made publicly available at https://github.com/aim-uofa/EvoIF.
SafeProtein: Red-Teaming Framework and Benchmark for Protein Foundation Models
Sep 03, 2025



Abstract:Proteins play crucial roles in almost all biological processes. The advancement of deep learning has greatly accelerated the development of protein foundation models, leading to significant successes in protein understanding and design. However, the lack of systematic red-teaming for these models has raised serious concerns about their potential misuse, such as generating proteins with biological safety risks. This paper introduces SafeProtein, the first red-teaming framework designed for protein foundation models to the best of our knowledge. SafeProtein combines multimodal prompt engineering and heuristic beam search to systematically design red-teaming methods and conduct tests on protein foundation models. We also curated SafeProtein-Bench, which includes a manually constructed red-teaming benchmark dataset and a comprehensive evaluation protocol. SafeProtein achieved continuous jailbreaks on state-of-the-art protein foundation models (up to 70% attack success rate for ESM3), revealing potential biological safety risks in current protein foundation models and providing insights for the development of robust security protection technologies for frontier models. The codes will be made publicly available at https://github.com/jigang-fan/SafeProtein.
Deciphering the unique dynamic activation pathway in a G protein-coupled receptor enables unveiling biased signaling and identifying cryptic allosteric sites in conformational intermediates
Apr 24, 2025



Abstract:Neurotensin receptor 1 (NTSR1), a member of the Class A G protein-coupled receptor superfamily, plays an important role in modulating dopaminergic neuronal activity and eliciting opioid-independent analgesia. Recent studies suggest that promoting \{beta}-arrestin-biased signaling in NTSR1 may diminish drugs of abuse, such as psychostimulants, thereby offering a potential avenue for treating human addiction-related disorders. In this study, we utilized a novel computational and experimental approach that combined nudged elastic band-based molecular dynamics simulations, Markov state models, temporal communication network analysis, site-directed mutagenesis, and conformational biosensors, to explore the intricate mechanisms underlying NTSR1 activation and biased signaling. Our study reveals a dynamic stepwise transition mechanism and activated transmission network associated with NTSR1 activation. It also yields valuable insights into the complex interplay between the unique polar network, non-conserved ion locks, and aromatic clusters in NTSR1 signaling. Moreover, we identified a cryptic allosteric site located in the intracellular region of the receptor that exists in an intermediate state within the activation pathway. Collectively, these findings contribute to a more profound understanding of NTSR1 activation and biased signaling at the atomic level, thereby providing a potential strategy for the development of NTSR1 allosteric modulators in the realm of G protein-coupled receptor biology, biophysics, and medicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge