Jian Zhan
A Comparative Review of RNA Language Models
May 14, 2025



Abstract:Given usefulness of protein language models (LMs) in structure and functional inference, RNA LMs have received increased attentions in the last few years. However, these RNA models are often not compared against the same standard. Here, we divided RNA LMs into three classes (pretrained on multiple RNA types (especially noncoding RNAs), specific-purpose RNAs, and LMs that unify RNA with DNA or proteins or both) and compared 13 RNA LMs along with 3 DNA and 1 protein LMs as controls in zero-shot prediction of RNA secondary structure and functional classification. Results shows that the models doing well on secondary structure prediction often perform worse in function classification or vice versa, suggesting that more balanced unsupervised training is needed.
Efficient Multi-Scale Attention Module with Cross-Spatial Learning
May 23, 2023



Abstract:Remarkable effectiveness of the channel or spatial attention mechanisms for producing more discernible feature representation are illustrated in various computer vision tasks. However, modeling the cross-channel relationships with channel dimensionality reduction may bring side effect in extracting deep visual representations. In this paper, a novel efficient multi-scale attention (EMA) module is proposed. Focusing on retaining the information on per channel and decreasing the computational overhead, we reshape the partly channels into the batch dimensions and group the channel dimensions into multiple sub-features which make the spatial semantic features well-distributed inside each feature group. Specifically, apart from encoding the global information to re-calibrate the channel-wise weight in each parallel branch, the output features of the two parallel branches are further aggregated by a cross-dimension interaction for capturing pixel-level pairwise relationship. We conduct extensive ablation studies and experiments on image classification and object detection tasks with popular benchmarks (e.g., CIFAR-100, ImageNet-1k, MS COCO and VisDrone2019) for evaluating its performance.
Autonomous Tissue Scanning under Free-Form Motion for Intraoperative Tissue Characterisation
May 22, 2020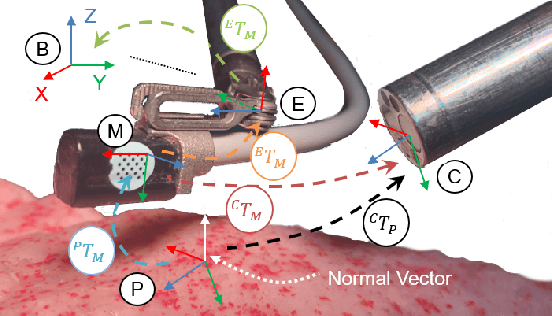
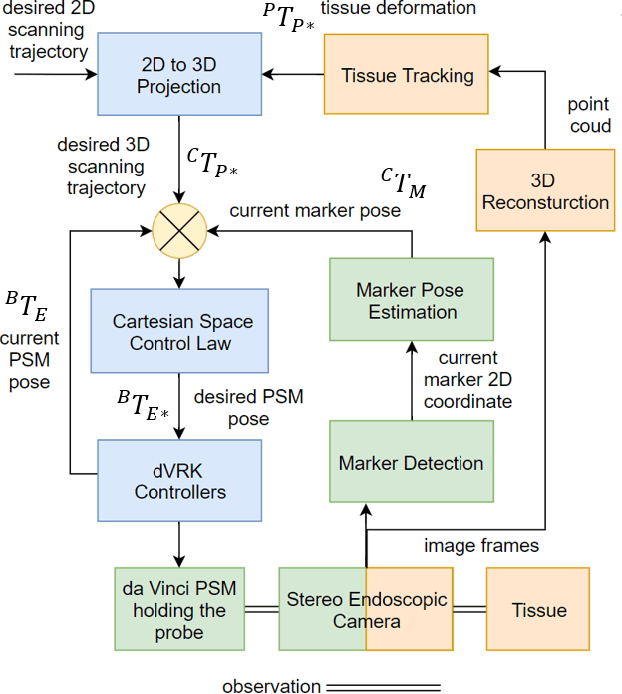
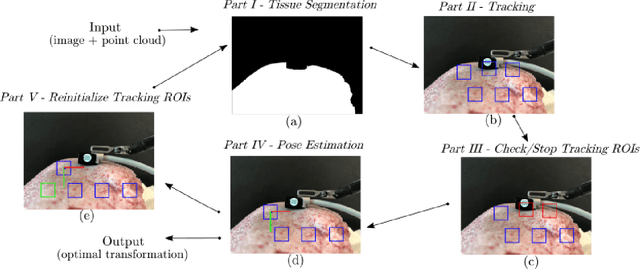
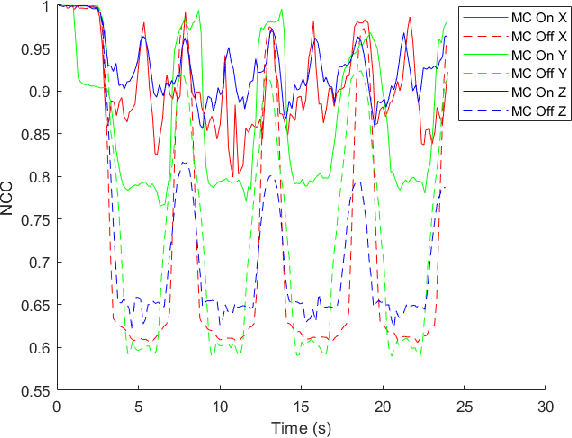
Abstract:In Minimally Invasive Surgery (MIS), tissue scanning with imaging probes is required for subsurface visualisation to characterise the state of the tissue. However, scanning of large tissue surfaces in the presence of deformation is a challenging task for the surgeon. Recently, robot-assisted local tissue scanning has been investigated for motion stabilisation of imaging probes to facilitate the capturing of good quality images and reduce the surgeon's cognitive load. Nonetheless, these approaches require the tissue surface to be static or deform with periodic motion. To eliminate these assumptions, we propose a visual servoing framework for autonomous tissue scanning, able to deal with free-form tissue deformation. The 3D structure of the surgical scene is recovered and a feature-based method is proposed to estimate the motion of the tissue in real-time. A desired scanning trajectory is manually defined on a reference frame and continuously updated using projective geometry to follow the tissue motion and control the movement of the robotic arm. The advantage of the proposed method is that it does not require the learning of the tissue motion prior to scanning and can deal with free-form deformation. We deployed this framework on the da Vinci surgical robot using the da Vinci Research Kit (dVRK) for Ultrasound tissue scanning. Since the framework does not rely on information from the Ultrasound data, it can be easily extended to other probe-based imaging modalities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge