Artificial Intelligence Assistance Significantly Improves Gleason Grading of Prostate Biopsies by Pathologists
Feb 11, 2020Wouter Bulten, Maschenka Balkenhol, Jean-Joël Awoumou Belinga, Américo Brilhante, Aslı Çakır, Xavier Farré, Katerina Geronatsiou, Vincent Molinié, Guilherme Pereira, Paromita Roy, Günter Saile, Paulo Salles, Ewout Schaafsma, Joëlle Tschui, Anne-Marie Vos, Hester van Boven, Robert Vink, Jeroen van der Laak, Christina Hulsbergen-van de Kaa, Geert Litjens
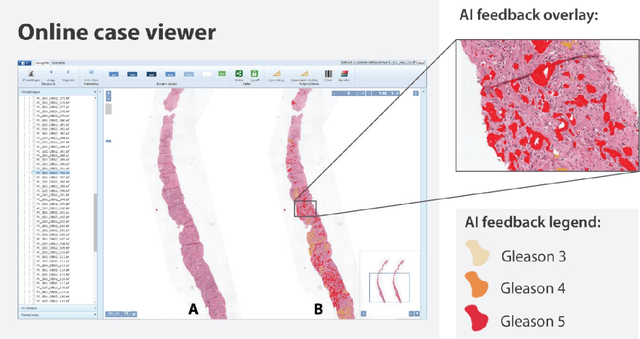
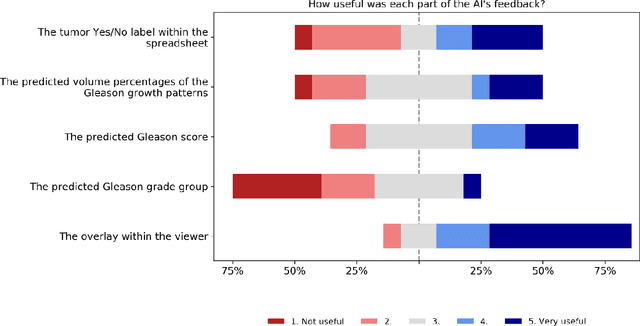
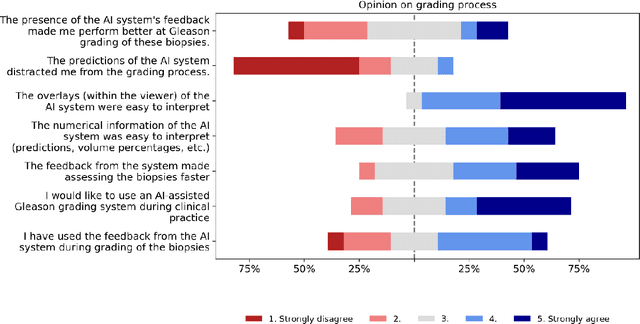
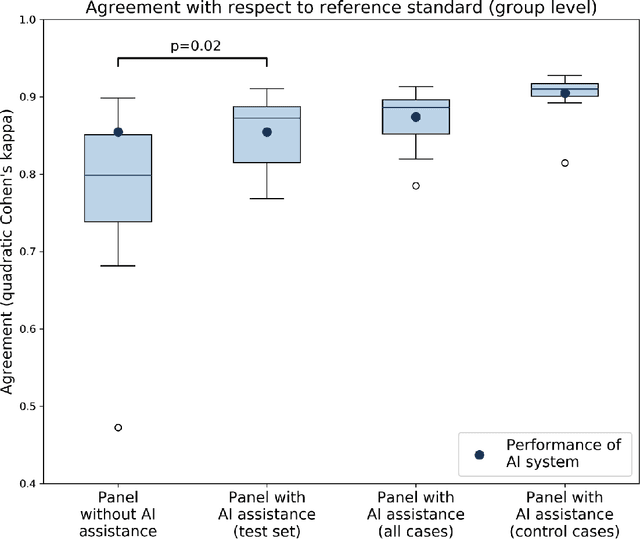
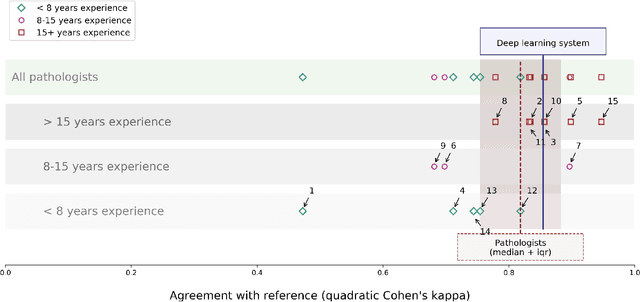
While the Gleason score is the most important prognostic marker for prostate cancer patients, it suffers from significant observer variability. Artificial Intelligence (AI) systems, based on deep learning, have proven to achieve pathologist-level performance at Gleason grading. However, the performance of such systems can degrade in the presence of artifacts, foreign tissue, or other anomalies. Pathologists integrating their expertise with feedback from an AI system could result in a synergy that outperforms both the individual pathologist and the system. Despite the hype around AI assistance, existing literature on this topic within the pathology domain is limited. We investigated the value of AI assistance for grading prostate biopsies. A panel of fourteen observers graded 160 biopsies with and without AI assistance. Using AI, the agreement of the panel with an expert reference standard significantly increased (quadratically weighted Cohen's kappa, 0.799 vs 0.872; p=0.018). Our results show the added value of AI systems for Gleason grading, but more importantly, show the benefits of pathologist-AI synergy.
Automated Gleason Grading of Prostate Biopsies using Deep Learning
Jul 18, 2019Wouter Bulten, Hans Pinckaers, Hester van Boven, Robert Vink, Thomas de Bel, Bram van Ginneken, Jeroen van der Laak, Christina Hulsbergen-van de Kaa, Geert Litjens
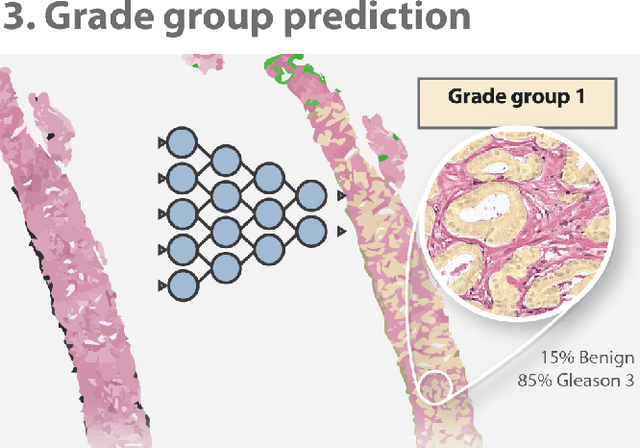
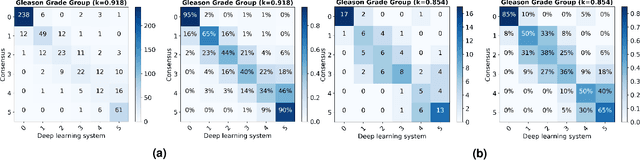
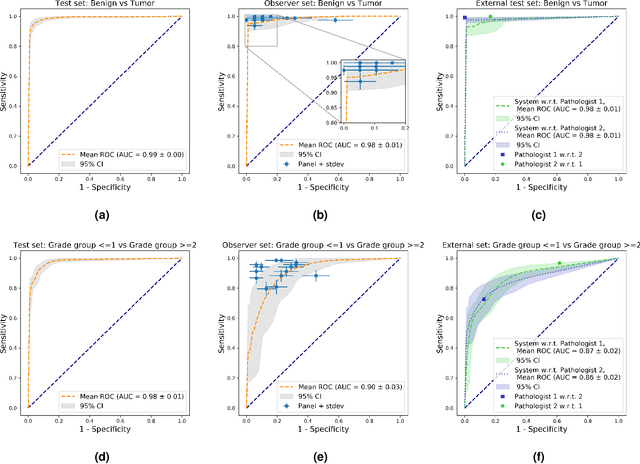
The Gleason score is the most important prognostic marker for prostate cancer patients but suffers from significant inter-observer variability. We developed a fully automated deep learning system to grade prostate biopsies. The system was developed using 5834 biopsies from 1243 patients. A semi-automatic labeling technique was used to circumvent the need for full manual annotation by pathologists. The developed system achieved a high agreement with the reference standard. In a separate observer experiment, the deep learning system outperformed 10 out of 15 pathologists. The system has the potential to improve prostate cancer prognostics by acting as a first or second reader.
Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology
Feb 18, 2019David Tellez, Geert Litjens, Peter Bandi, Wouter Bulten, John-Melle Bokhorst, Francesco Ciompi, Jeroen van der Laak
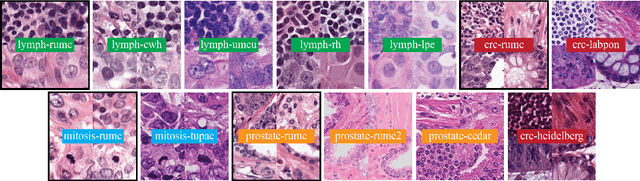
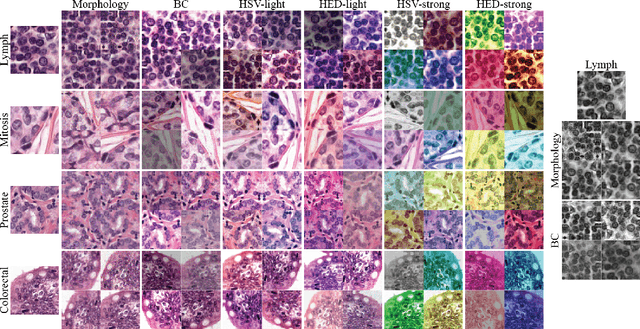
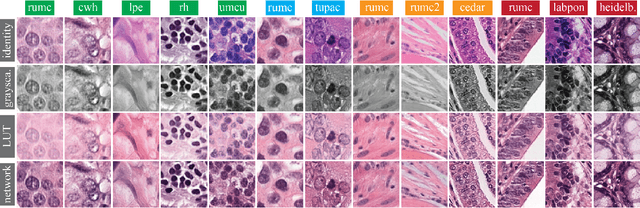
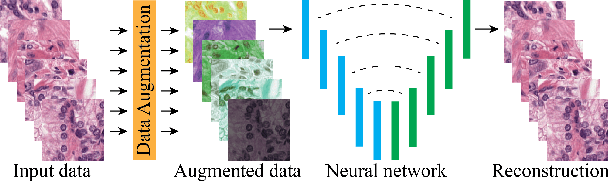
Stain variation is a phenomenon observed when distinct pathology laboratories stain tissue slides that exhibit similar but not identical color appearance. Due to this color shift between laboratories, convolutional neural networks (CNNs) trained with images from one lab often underperform on unseen images from the other lab. Several techniques have been proposed to reduce the generalization error, mainly grouped into two categories: stain color augmentation and stain color normalization. The former simulates a wide variety of realistic stain variations during training, producing stain-invariant CNNs. The latter aims to match training and test color distributions in order to reduce stain variation. For the first time, we compared some of these techniques and quantified their effect on CNN classification performance using a heterogeneous dataset of hematoxylin and eosin histopathology images from 4 organs and 9 pathology laboratories. Additionally, we propose a novel unsupervised method to perform stain color normalization using a neural network. Based on our experimental results, we provide practical guidelines on how to use stain color augmentation and stain color normalization in future computational pathology applications.
Neural Image Compression for Gigapixel Histopathology Image Analysis
Nov 07, 2018David Tellez, Geert Litjens, Jeroen van der Laak, Francesco Ciompi
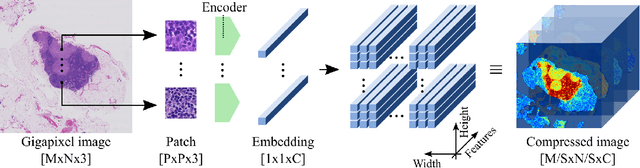
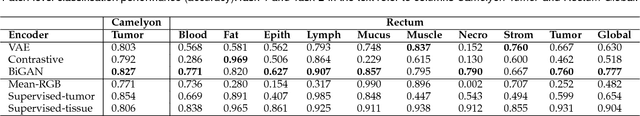
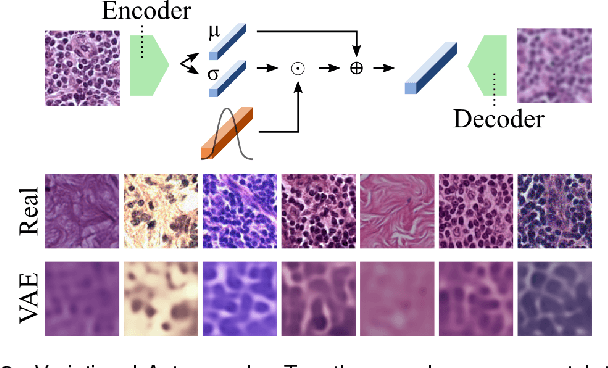
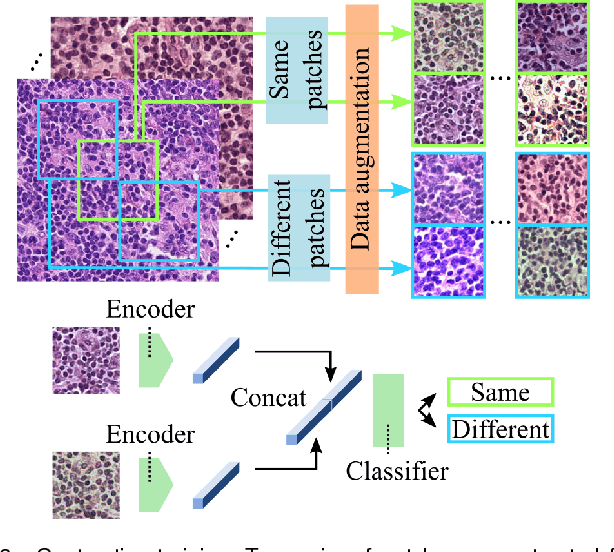
We present Neural Image Compression (NIC), a method to reduce the size of gigapixel images by mapping them to a compact latent space using neural networks. We show that this compression allows us to train convolutional neural networks on histopathology whole-slide images end-to-end using weak image-level labels.
Whole-Slide Mitosis Detection in H&E Breast Histology Using PHH3 as a Reference to Train Distilled Stain-Invariant Convolutional Networks
Aug 17, 2018David Tellez, Maschenka Balkenhol, Irene Otte-Holler, Rob van de Loo, Rob Vogels, Peter Bult, Carla Wauters, Willem Vreuls, Suzanne Mol, Nico Karssemeijer, Geert Litjens, Jeroen van der Laak, Francesco Ciompi
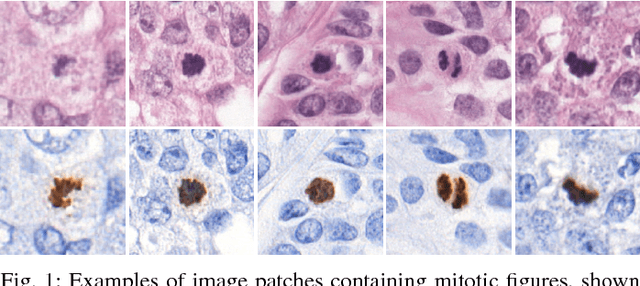
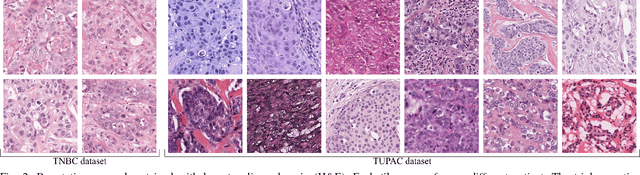
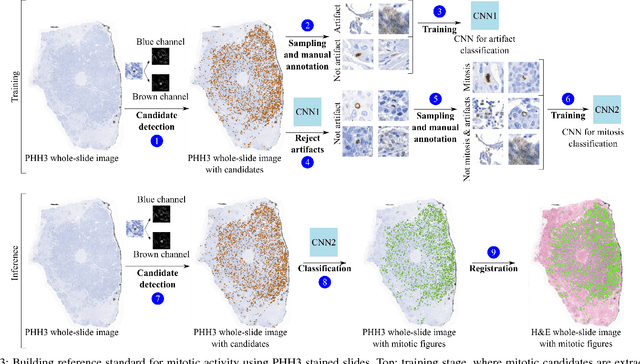
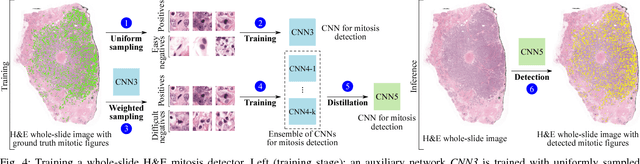
Manual counting of mitotic tumor cells in tissue sections constitutes one of the strongest prognostic markers for breast cancer. This procedure, however, is time-consuming and error-prone. We developed a method to automatically detect mitotic figures in breast cancer tissue sections based on convolutional neural networks (CNNs). Application of CNNs to hematoxylin and eosin (H&E) stained histological tissue sections is hampered by: (1) noisy and expensive reference standards established by pathologists, (2) lack of generalization due to staining variation across laboratories, and (3) high computational requirements needed to process gigapixel whole-slide images (WSIs). In this paper, we present a method to train and evaluate CNNs to specifically solve these issues in the context of mitosis detection in breast cancer WSIs. First, by combining image analysis of mitotic activity in phosphohistone-H3 (PHH3) restained slides and registration, we built a reference standard for mitosis detection in entire H&E WSIs requiring minimal manual annotation effort. Second, we designed a data augmentation strategy that creates diverse and realistic H&E stain variations by modifying the hematoxylin and eosin color channels directly. Using it during training combined with network ensembling resulted in a stain invariant mitosis detector. Third, we applied knowledge distillation to reduce the computational requirements of the mitosis detection ensemble with a negligible loss of performance. The system was trained in a single-center cohort and evaluated in an independent multicenter cohort from The Cancer Genome Atlas on the three tasks of the Tumor Proliferation Assessment Challenge (TUPAC). We obtained a performance within the top-3 best methods for most of the tasks of the challenge.
Epithelium segmentation using deep learning in H&E-stained prostate specimens with immunohistochemistry as reference standard
Aug 17, 2018Wouter Bulten, Péter Bándi, Jeffrey Hoven, Rob van de Loo, Johannes Lotz, Nick Weiss, Jeroen van der Laak, Bram van Ginneken, Christina Hulsbergen-van de Kaa, Geert Litjens
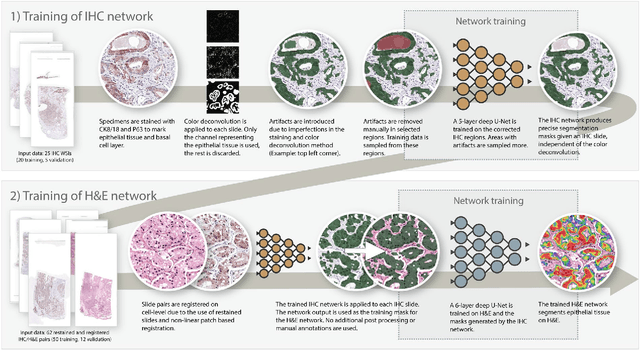
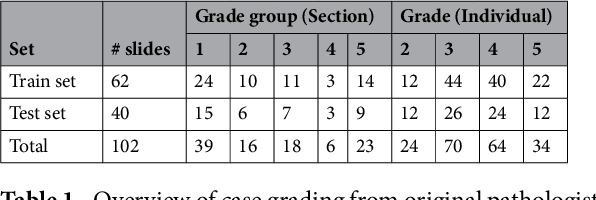
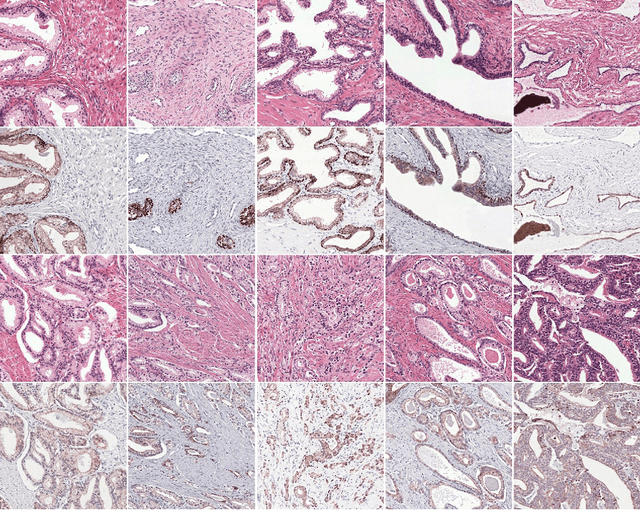
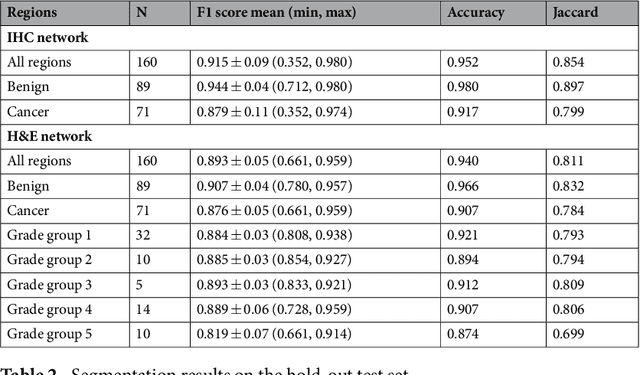
Prostate cancer (PCa) is graded by pathologists by examining the architectural pattern of cancerous epithelial tissue on hematoxylin and eosin (H&E) stained slides. Given the importance of gland morphology, automatically differentiating between glandular epithelial tissue and other tissues is an important prerequisite for the development of automated methods for detecting PCa. We propose a new method, using deep learning, for automatically segmenting epithelial tissue in digitized prostatectomy slides. We employed immunohistochemistry (IHC) to render the ground truth less subjective and more precise compared to manual outlining on H&E slides, especially in areas with high-grade and poorly differentiated PCa. Our dataset consisted of 102 tissue blocks, including both low and high grade PCa. From each block a single new section was cut, stained with H&E, scanned, restained using P63 and CK8/18 to highlight the epithelial structure, and scanned again. The H&E slides were co-registered to the IHC slides. On a subset of the IHC slides we applied color deconvolution, corrected stain errors manually, and trained a U-Net to perform segmentation of epithelial structures. Whole-slide segmentation masks generated by the IHC U-Net were used to train a second U-Net on H&E. Our system makes precise cell-level segmentations and segments both intact glands as well as individual (tumor) epithelial cells. We achieved an F1-score of 0.895 on a hold-out test set and 0.827 on an external reference set from a different center. We envision this segmentation as being the first part of a fully automated prostate cancer detection and grading pipeline.
The importance of stain normalization in colorectal tissue classification with convolutional networks
May 23, 2017Francesco Ciompi, Oscar Geessink, Babak Ehteshami Bejnordi, Gabriel Silva de Souza, Alexi Baidoshvili, Geert Litjens, Bram van Ginneken, Iris Nagtegaal, Jeroen van der Laak
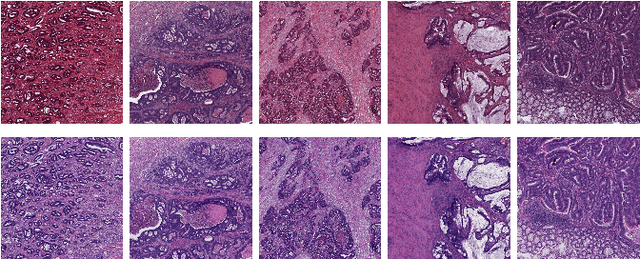

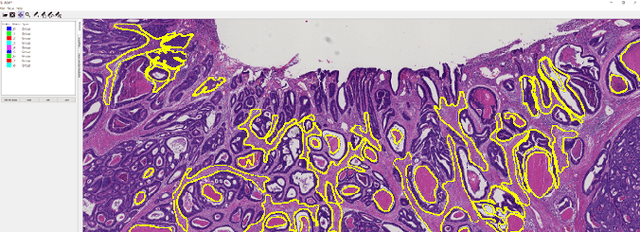
The development of reliable imaging biomarkers for the analysis of colorectal cancer (CRC) in hematoxylin and eosin (H&E) stained histopathology images requires an accurate and reproducible classification of the main tissue components in the image. In this paper, we propose a system for CRC tissue classification based on convolutional networks (ConvNets). We investigate the importance of stain normalization in tissue classification of CRC tissue samples in H&E-stained images. Furthermore, we report the performance of ConvNets on a cohort of rectal cancer samples and on an independent publicly available dataset of colorectal H&E images.
Context-aware stacked convolutional neural networks for classification of breast carcinomas in whole-slide histopathology images
May 10, 2017Babak Ehteshami Bejnordi, Guido Zuidhof, Maschenka Balkenhol, Meyke Hermsen, Peter Bult, Bram van Ginneken, Nico Karssemeijer, Geert Litjens, Jeroen van der Laak
Automated classification of histopathological whole-slide images (WSI) of breast tissue requires analysis at very high resolutions with a large contextual area. In this paper, we present context-aware stacked convolutional neural networks (CNN) for classification of breast WSIs into normal/benign, ductal carcinoma in situ (DCIS), and invasive ductal carcinoma (IDC). We first train a CNN using high pixel resolution patches to capture cellular level information. The feature responses generated by this model are then fed as input to a second CNN, stacked on top of the first. Training of this stacked architecture with large input patches enables learning of fine-grained (cellular) details and global interdependence of tissue structures. Our system is trained and evaluated on a dataset containing 221 WSIs of H&E stained breast tissue specimens. The system achieves an AUC of 0.962 for the binary classification of non-malignant and malignant slides and obtains a three class accuracy of 81.3% for classification of WSIs into normal/benign, DCIS, and IDC, demonstrating its potentials for routine diagnostics.
Comparison of Different Methods for Tissue Segmentation in Histopathological Whole-Slide Images
Apr 03, 2017Péter Bándi, Rob van de Loo, Milad Intezar, Daan Geijs, Francesco Ciompi, Bram van Ginneken, Jeroen van der Laak, Geert Litjens
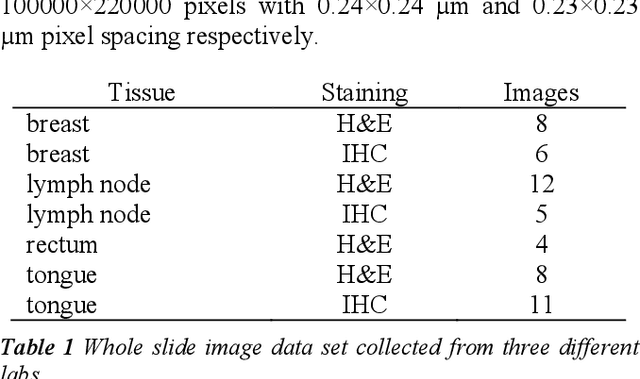
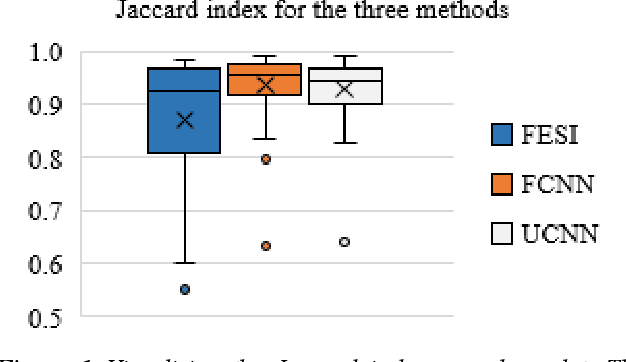
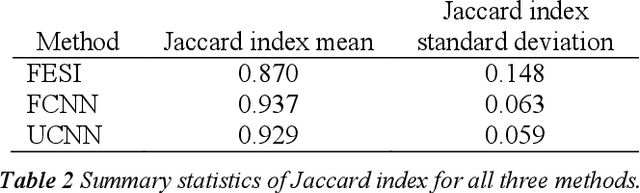
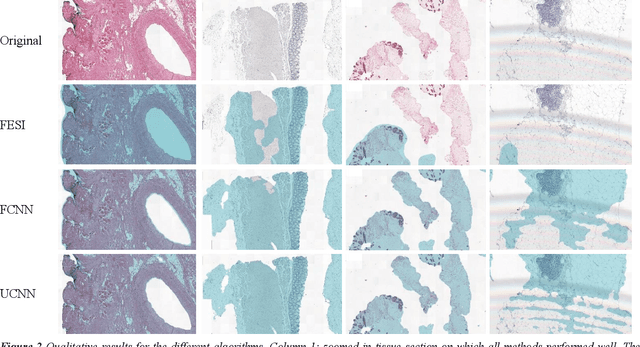
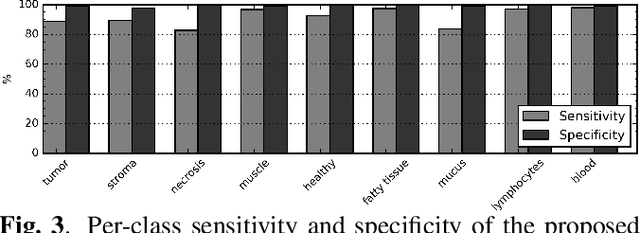
Tissue segmentation is an important pre-requisite for efficient and accurate diagnostics in digital pathology. However, it is well known that whole-slide scanners can fail in detecting all tissue regions, for example due to the tissue type, or due to weak staining because their tissue detection algorithms are not robust enough. In this paper, we introduce two different convolutional neural network architectures for whole slide image segmentation to accurately identify the tissue sections. We also compare the algorithms to a published traditional method. We collected 54 whole slide images with differing stains and tissue types from three laboratories to validate our algorithms. We show that while the two methods do not differ significantly they outperform their traditional counterpart (Jaccard index of 0.937 and 0.929 vs. 0.870, p < 0.01).
Deep learning-based assessment of tumor-associated stroma for diagnosing breast cancer in histopathology images
Feb 19, 2017Babak Ehteshami Bejnordi, Jimmy Linz, Ben Glass, Maeve Mullooly, Gretchen L Gierach, Mark E Sherman, Nico Karssemeijer, Jeroen van der Laak, Andrew H Beck
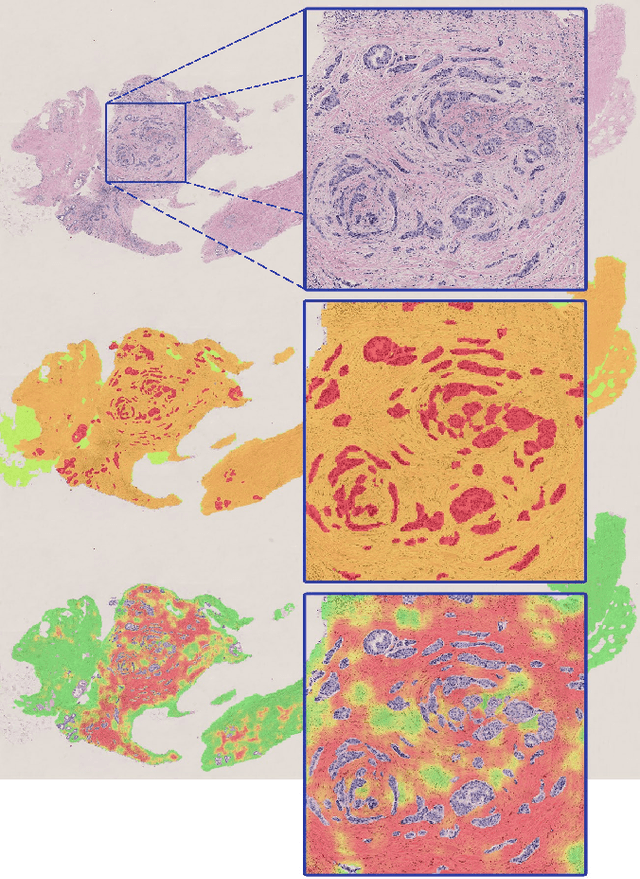
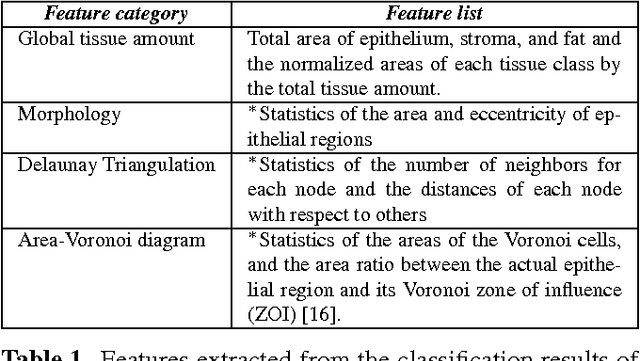
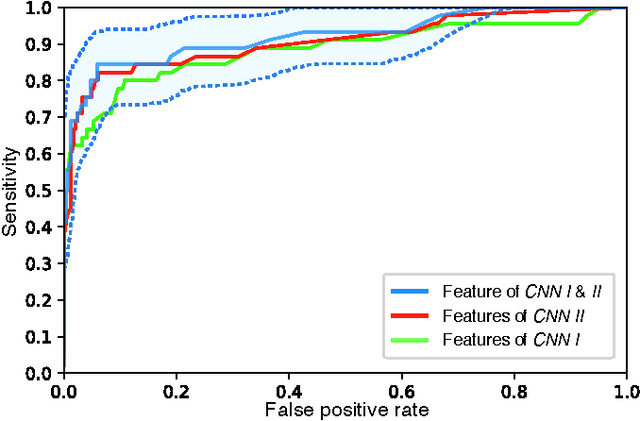
Diagnosis of breast carcinomas has so far been limited to the morphological interpretation of epithelial cells and the assessment of epithelial tissue architecture. Consequently, most of the automated systems have focused on characterizing the epithelial regions of the breast to detect cancer. In this paper, we propose a system for classification of hematoxylin and eosin (H&E) stained breast specimens based on convolutional neural networks that primarily targets the assessment of tumor-associated stroma to diagnose breast cancer patients. We evaluate the performance of our proposed system using a large cohort containing 646 breast tissue biopsies. Our evaluations show that the proposed system achieves an area under ROC of 0.92, demonstrating the discriminative power of previously neglected tumor-associated stroma as a diagnostic biomarker.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge