Ignacio Arganda-Carreras
Deep Learning for Accurate Vision-based Catch Composition in Tropical Tuna Purse Seiners
Nov 19, 2025



Abstract:Purse seiners play a crucial role in tuna fishing, as approximately 69% of the world's tropical tuna is caught using this gear. All tuna Regional Fisheries Management Organizations have established minimum standards to use electronic monitoring (EM) in fisheries in addition to traditional observers. The EM systems produce a massive amount of video data that human analysts must process. Integrating artificial intelligence (AI) into their workflow can decrease that workload and improve the accuracy of the reports. However, species identification still poses significant challenges for AI, as achieving balanced performance across all species requires appropriate training data. Here, we quantify the difficulty experts face to distinguish bigeye tuna (BET, Thunnus Obesus) from yellowfin tuna (YFT, Thunnus Albacares) using images captured by EM systems. We found inter-expert agreements of 42.9% $\pm$ 35.6% for BET and 57.1% $\pm$ 35.6% for YFT. We then present a multi-stage pipeline to estimate the species composition of the catches using a reliable ground-truth dataset based on identifications made by observers on board. Three segmentation approaches are compared: Mask R-CNN, a combination of DINOv2 with SAM2, and a integration of YOLOv9 with SAM2. We found that the latest performs the best, with a validation mean average precision of 0.66 $\pm$ 0.03 and a recall of 0.88 $\pm$ 0.03. Segmented individuals are tracked using ByteTrack. For classification, we evaluate a standard multiclass classification model and a hierarchical approach, finding a superior generalization by the hierarchical. All our models were cross-validated during training and tested on fishing operations with fully known catch composition. Combining YOLOv9-SAM2 with the hierarchical classification produced the best estimations, with 84.8% of the individuals being segmented and classified with a mean average error of 4.5%.
A Fully Interpretable Statistical Approach for Roadside LiDAR Background Subtraction
Oct 25, 2025



Abstract:We present a fully interpretable and flexible statistical method for background subtraction in roadside LiDAR data, aimed at enhancing infrastructure-based perception in automated driving. Our approach introduces both a Gaussian distribution grid (GDG), which models the spatial statistics of the background using background-only scans, and a filtering algorithm that uses this representation to classify LiDAR points as foreground or background. The method supports diverse LiDAR types, including multiline 360 degree and micro-electro-mechanical systems (MEMS) sensors, and adapts to various configurations. Evaluated on the publicly available RCooper dataset, it outperforms state-of-the-art techniques in accuracy and flexibility, even with minimal background data. Its efficient implementation ensures reliable performance on low-resource hardware, enabling scalable real-world deployment.
Learning Gaze-aware Compositional GAN
May 31, 2024



Abstract:Gaze-annotated facial data is crucial for training deep neural networks (DNNs) for gaze estimation. However, obtaining these data is labor-intensive and requires specialized equipment due to the challenge of accurately annotating the gaze direction of a subject. In this work, we present a generative framework to create annotated gaze data by leveraging the benefits of labeled and unlabeled data sources. We propose a Gaze-aware Compositional GAN that learns to generate annotated facial images from a limited labeled dataset. Then we transfer this model to an unlabeled data domain to take advantage of the diversity it provides. Experiments demonstrate our approach's effectiveness in generating within-domain image augmentations in the ETH-XGaze dataset and cross-domain augmentations in the CelebAMask-HQ dataset domain for gaze estimation DNN training. We also show additional applications of our work, which include facial image editing and gaze redirection.
* Accepted by ETRA 2024 as Full paper, and as journal paper in Proceedings of the ACM on Computer Graphics and Interactive Techniques
Instance Segmentation of Unlabeled Modalities via Cyclic Segmentation GAN
Apr 06, 2022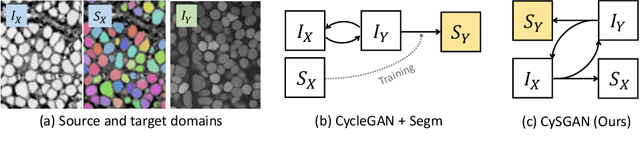
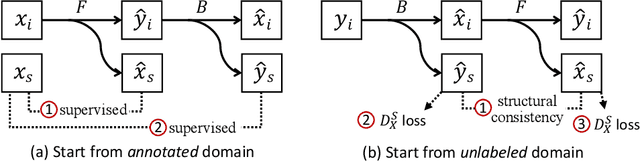
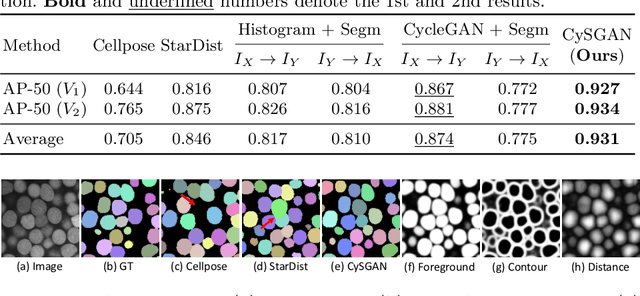
Abstract:Instance segmentation for unlabeled imaging modalities is a challenging but essential task as collecting expert annotation can be expensive and time-consuming. Existing works segment a new modality by either deploying a pre-trained model optimized on diverse training data or conducting domain translation and image segmentation as two independent steps. In this work, we propose a novel Cyclic Segmentation Generative Adversarial Network (CySGAN) that conducts image translation and instance segmentation jointly using a unified framework. Besides the CycleGAN losses for image translation and supervised losses for the annotated source domain, we introduce additional self-supervised and segmentation-based adversarial objectives to improve the model performance by leveraging unlabeled target domain images. We benchmark our approach on the task of 3D neuronal nuclei segmentation with annotated electron microscopy (EM) images and unlabeled expansion microscopy (ExM) data. Our CySGAN outperforms both pretrained generalist models and the baselines that sequentially conduct image translation and segmentation. Our implementation and the newly collected, densely annotated ExM nuclei dataset, named NucExM, are available at https://connectomics-bazaar.github.io/proj/CySGAN/index.html.
Deep learning based domain adaptation for mitochondria segmentation on EM volumes
Feb 22, 2022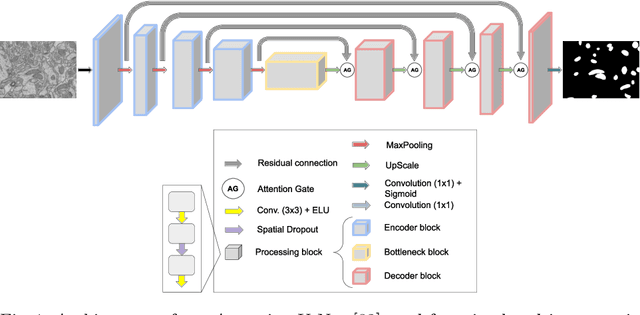

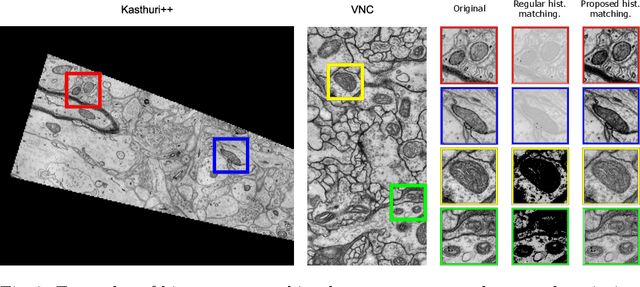
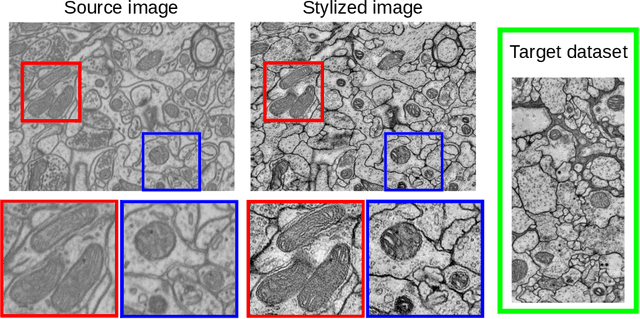
Abstract:Accurate segmentation of electron microscopy (EM) volumes of the brain is essential to characterize neuronal structures at a cell or organelle level. While supervised deep learning methods have led to major breakthroughs in that direction during the past years, they usually require large amounts of annotated data to be trained, and perform poorly on other data acquired under similar experimental and imaging conditions. This is a problem known as domain adaptation, since models that learned from a sample distribution (or source domain) struggle to maintain their performance on samples extracted from a different distribution or target domain. In this work, we address the complex case of deep learning based domain adaptation for mitochondria segmentation across EM datasets from different tissues and species. We present three unsupervised domain adaptation strategies to improve mitochondria segmentation in the target domain based on (1) state-of-the-art style transfer between images of both domains; (2) self-supervised learning to pre-train a model using unlabeled source and target images, and then fine-tune it only with the source labels; and (3) multi-task neural network architectures trained end-to-end with both labeled and unlabeled images. Additionally, we propose a new training stopping criterion based on morphological priors obtained exclusively in the source domain. We carried out all possible cross-dataset experiments using three publicly available EM datasets. We evaluated our proposed strategies on the mitochondria semantic labels predicted on the target datasets. The methods introduced here outperform the baseline methods and compare favorably to the state of the art. In the absence of validation labels, monitoring our proposed morphology-based metric is an intuitive and effective way to stop the training process and select in average optimal models.
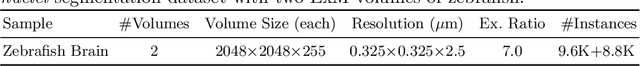
NucMM Dataset: 3D Neuronal Nuclei Instance Segmentation at Sub-Cubic Millimeter Scale
Jul 13, 2021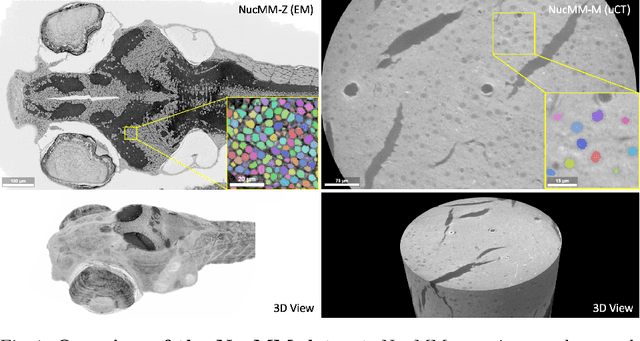

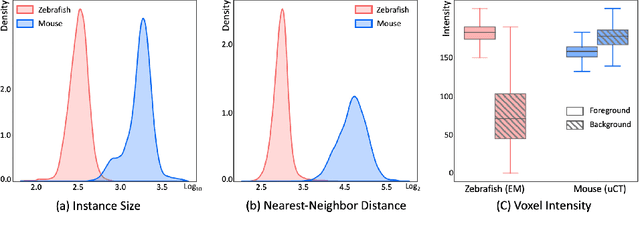
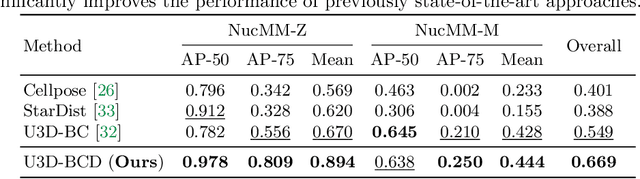
Abstract:Segmenting 3D cell nuclei from microscopy image volumes is critical for biological and clinical analysis, enabling the study of cellular expression patterns and cell lineages. However, current datasets for neuronal nuclei usually contain volumes smaller than $10^{\text{-}3}\ mm^3$ with fewer than 500 instances per volume, unable to reveal the complexity in large brain regions and restrict the investigation of neuronal structures. In this paper, we have pushed the task forward to the sub-cubic millimeter scale and curated the NucMM dataset with two fully annotated volumes: one $0.1\ mm^3$ electron microscopy (EM) volume containing nearly the entire zebrafish brain with around 170,000 nuclei; and one $0.25\ mm^3$ micro-CT (uCT) volume containing part of a mouse visual cortex with about 7,000 nuclei. With two imaging modalities and significantly increased volume size and instance numbers, we discover a great diversity of neuronal nuclei in appearance and density, introducing new challenges to the field. We also perform a statistical analysis to illustrate those challenges quantitatively. To tackle the challenges, we propose a novel hybrid-representation learning model that combines the merits of foreground mask, contour map, and signed distance transform to produce high-quality 3D masks. The benchmark comparisons on the NucMM dataset show that our proposed method significantly outperforms state-of-the-art nuclei segmentation approaches. Code and data are available at https://connectomics-bazaar.github.io/proj/nucMM/index.html.
AxonEM Dataset: 3D Axon Instance Segmentation of Brain Cortical Regions
Jul 12, 2021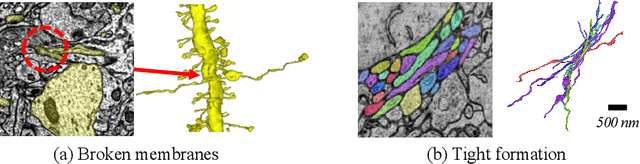

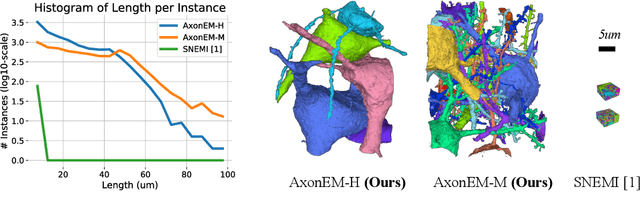
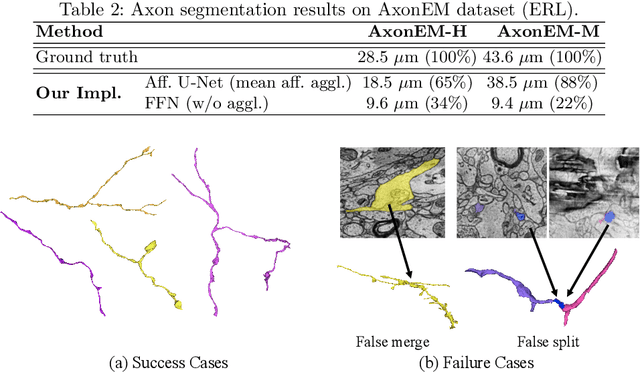
Abstract:Electron microscopy (EM) enables the reconstruction of neural circuits at the level of individual synapses, which has been transformative for scientific discoveries. However, due to the complex morphology, an accurate reconstruction of cortical axons has become a major challenge. Worse still, there is no publicly available large-scale EM dataset from the cortex that provides dense ground truth segmentation for axons, making it difficult to develop and evaluate large-scale axon reconstruction methods. To address this, we introduce the AxonEM dataset, which consists of two 30x30x30 um^3 EM image volumes from the human and mouse cortex, respectively. We thoroughly proofread over 18,000 axon instances to provide dense 3D axon instance segmentation, enabling large-scale evaluation of axon reconstruction methods. In addition, we densely annotate nine ground truth subvolumes for training, per each data volume. With this, we reproduce two published state-of-the-art methods and provide their evaluation results as a baseline. We publicly release our code and data at https://connectomics-bazaar.github.io/proj/AxonEM/index.html to foster the development of advanced methods.
Stable deep neural network architectures for mitochondria segmentation on electron microscopy volumes
Apr 08, 2021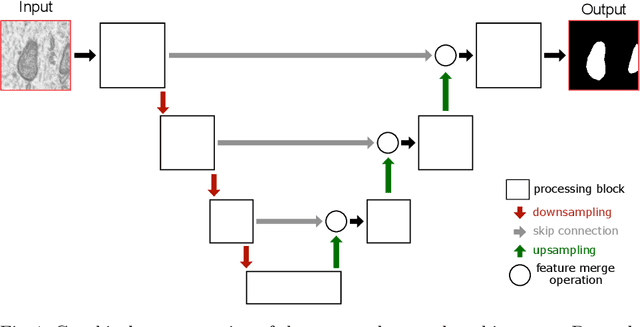
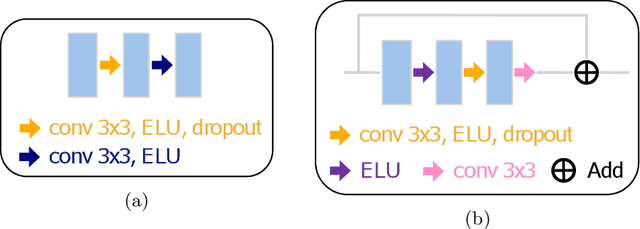
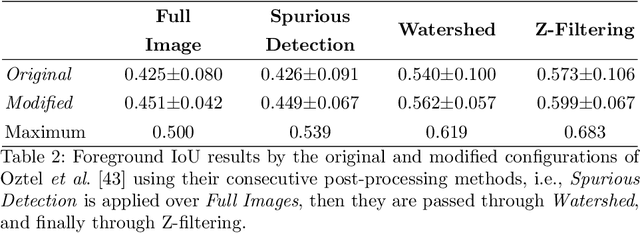
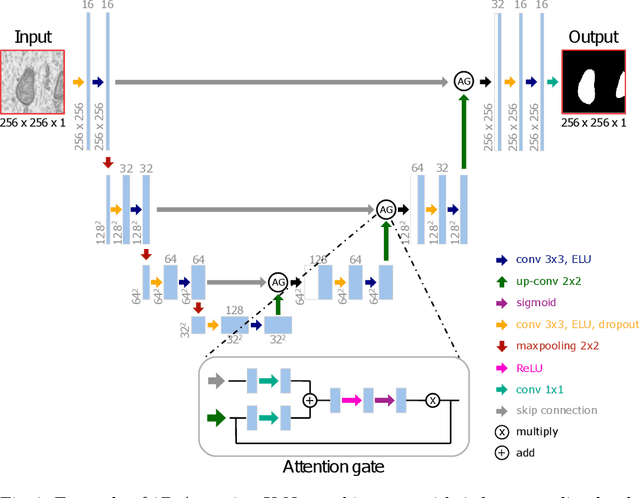
Abstract:Electron microscopy (EM) allows the identification of intracellular organelles such as mitochondria, providing insights for clinical and scientific studies. In recent years, a number of novel deep learning architectures have been published reporting superior performance, or even human-level accuracy, compared to previous approaches on public mitochondria segmentation datasets. Unfortunately, many of these publications do not make neither the code nor the full training details public to support the results obtained, leading to reproducibility issues and dubious model comparisons. For that reason, and following a recent code of best practices for reporting experimental results, we present an extensive study of the state-of-the-art deep learning architectures for the segmentation of mitochondria on EM volumes, and evaluate the impact in performance of different variations of 2D and 3D U-Net-like models for this task. To better understand the contribution of each component, a common set of pre- and post-processing operations has been implemented and tested with each approach. Moreover, an exhaustive sweep of hyperparameters values for all architectures have been performed and each configuration has been run multiple times to report the mean and standard deviation values of the evaluation metrics. Using this methodology, we found very stable architectures and hyperparameter configurations that consistently obtain state-of-the-art results in the well-known EPFL Hippocampus mitochondria segmentation dataset. Furthermore, we have benchmarked our proposed models on two other available datasets, Lucchi++ and Kasthuri++, where they outperform all previous works. The code derived from this research and its documentation are publicly available.
Inferring spatial relations from textual descriptions of images
Feb 01, 2021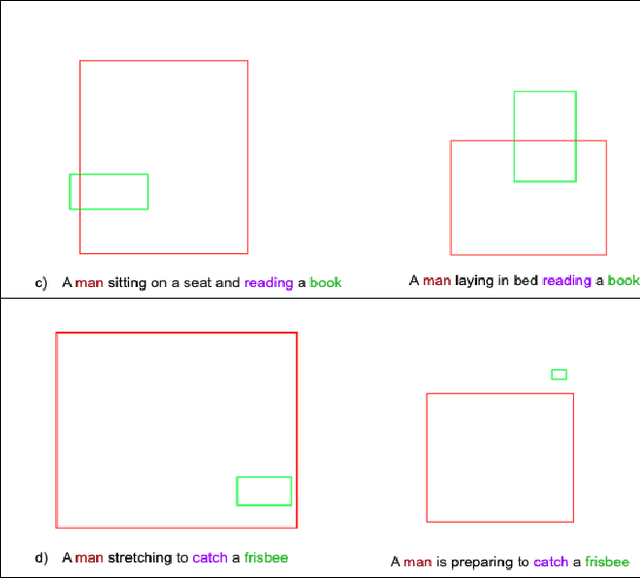



Abstract:Generating an image from its textual description requires both a certain level of language understanding and common sense knowledge about the spatial relations of the physical entities being described. In this work, we focus on inferring the spatial relation between entities, a key step in the process of composing scenes based on text. More specifically, given a caption containing a mention to a subject and the location and size of the bounding box of that subject, our goal is to predict the location and size of an object mentioned in the caption. Previous work did not use the caption text information, but a manually provided relation holding between the subject and the object. In fact, the used evaluation datasets contain manually annotated ontological triplets but no captions, making the exercise unrealistic: a manual step was required; and systems did not leverage the richer information in captions. Here we present a system that uses the full caption, and Relations in Captions (REC-COCO), a dataset derived from MS-COCO which allows to evaluate spatial relation inference from captions directly. Our experiments show that: (1) it is possible to infer the size and location of an object with respect to a given subject directly from the caption; (2) the use of full text allows to place the object better than using a manually annotated relation. Our work paves the way for systems that, given a caption, decide which entities need to be depicted and their respective location and sizes, in order to then generate the final image.
Deep Learning on Chest X-ray Images to Detect and Evaluate Pneumonia Cases at the Era of COVID-19
Apr 05, 2020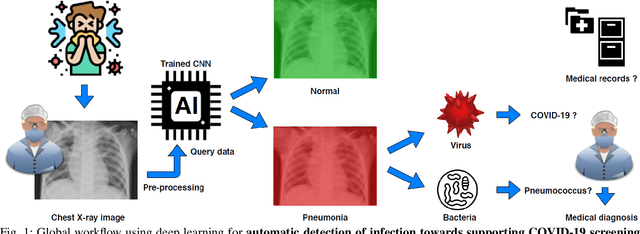
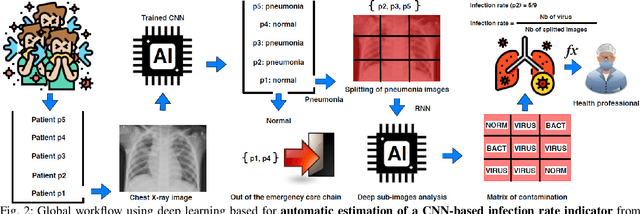
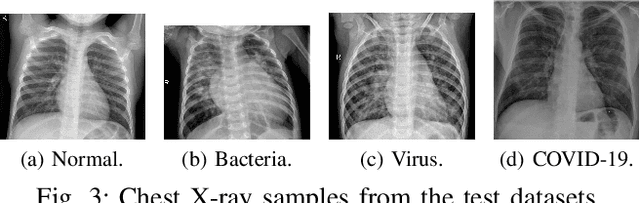
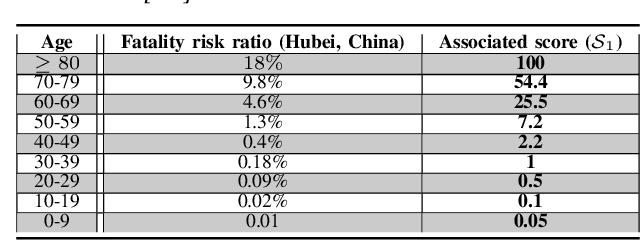
Abstract:Coronavirus disease 2019 (COVID-19) is an infectious disease with first symptoms similar to the flu. COVID-19 appeared first in China and very quickly spreads to the rest of the world, causing then the 2019-20 coronavirus pandemic. In many cases, this disease causes pneumonia. Since pulmonary infections can be observed through radiography images, this paper investigates deep learning methods for automatically analyzing query chest X-ray images with the hope to bring precision tools to health professionals towards screening the COVID-19 and diagnosing confirmed patients. In this context, training datasets, deep learning architectures and analysis strategies have been experimented from publicly open sets of chest X-ray images. Tailored deep learning models are proposed to detect pneumonia infection cases, notably viral cases. It is assumed that viral pneumonia cases detected during an epidemic COVID-19 context have a high probability to presume COVID-19 infections. Moreover, easy-to-apply health indicators are proposed for estimating infection status and predicting patient status from the detected pneumonia cases. Experimental results show possibilities of training deep learning models over publicly open sets of chest X-ray images towards screening viral pneumonia. Chest X-ray test images of COVID-19 infected patients are successfully diagnosed through detection models retained for their performances. The efficiency of proposed health indicators is highlighted through simulated scenarios of patients presenting infections and health problems by combining real and synthetic health data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge